
PANK1 associates with cancer metabolism and immune infiltration in clear cell renal cell carcinoma: a retrospective prognostic study based on the TCGA database
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common histological subtype of renal cell carcinoma and accounts for nearly 75% of all cases (1). For early stage ccRCC, surgical excision is the best treatment approach. However, despite substantial advances in ccRCC-targeted therapies, such as tyrosine kinase or mammalian target of rapamycin (mTOR) inhibitors, the clinical prognosis of patients with advanced and metastatic ccRCC remains unsatisfactory (2). Additionally, ccRCC is reported to be a highly immune-infiltrated tumor, and immunotherapy-based combination therapeutics have been shown to have increased clinical benefits in terms of their efficacy and the overall survival (OS) of patients with metastatic RCC (3,4). Recently, the general metabolism mitochondrial functions of RCC were reported to be correlated with clinical prognosis (5,6); however, the mechanisms of this correlation are not clear. Thus, investigating the mechanisms that link metabolism and immune infiltration is important for the pathogenesis and treatment of ccRCC.
Previous studies concentrate on the biological and clinical markers associated with the prognosis of ccRCC (7). Petitprez et al. reviewed a total of 341 distinct markers and 13 multiple-marker models for ccRCC, of which the main ones are distributed in cell cycle, angiogenesis, hypoxia, and immune response (8). Recent study indicates that ccRCC is regarded as a metabolic disease (9). However, still few study concentrated on the metabolic markers and clinical prognosis of ccRCC (10). Therefore, identifying the key metabolic markers associated with the clinical prognosis of ccRCC is important for further understanding of ccRCC.
Pantothenate kinase-1 (PANK1) is a member of the PanK family, and is involved in the biosynthesis of coenzyme A (CoA) (11). CoA has been shown to be an essential and prevalent cofactor that is essential in more than a hundred diverse metabolic reactions, including the tricarboxylic acid cycle, synthesis of fatty acids and neurotransmitters, activation of immune functions, and repair of connective tissues (12). The PanK family of proteins are responsible for catalyzing the first and rate-limiting step of CoA synthesis (the phosphorylation of the precursor pantothenate), and thus controlling the cellular content of CoA (13). The CoA levels, which are used to meet the energy demands as an alternative fuel source in tissues, change under metabolic stress. In pathological conditions, increase of intracellular CoA in the liver is required to guarantee the efficient conversion of fatty acids and amino acids in store to energy. Thus, PANK1 has been shown to play important roles in the metabolism regulation of glucose and lipids, and have promising effects in interventions of metabolism-related disorders (14).
However, little research has been conducted on whether PANK1 participates in cancer pathogenesis. Zi et al. reported that PANK1 inhibits hepatocellular carcinoma progression by modulating the Wnt/β-catenin signaling (11). Additionally, PANK1 has been reported to modulate cell cycle progression via the downregulation of cyclin dependent kinase 6 and p130 proteins (15). A recent bioinformatic analysis revealed that PANK1 is an important differentially expressed gene between normal and tumor tissues (16); however, the role of PANK1 in ccRCC has not yet been systematically investigated, especially its role in the metabolic regulation and clinical prognosis. In this study, we used data from The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) cohort to study the correlations between PANK1 expression and the clinical prognosis of ccRCC patients. The effects of PANK1 on cell viability and cell metabolism were also detected using ccRCC cell lines. Our data revealed the important role of PANK1 in ccRCC and its relationship to tumor metabolism and immunity. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1488/rc).
Methods
Study design and research procedures
This study is a retrospective prognostic study analyzing the expression difference of PANK1 between tumor and adjacent normal tissues, correlations with the clinical factors, overall and disease-specific survival analyses using the TCGA-KIRC data. Moreover, receiver operator characteristic curve (ROC), time-dependent ROC and nomogram model incorporating key clinical factors and PANK1 expression patterns were built. In addition, promoter methylation status of PANK1 and the correlations with key clinical factors were investigated. Then, signaling pathways and functional protein networks were clustered, while the associations between PANK1 expression, immune cell infiltrations were presented. At last, in vitro cell experiments were conducted to further validate the results from the clinical analyses. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene expression profile analysis
PANK1 messenger ribonucleic acid (mRNA) levels in several cancer types were analyzed using the Tumor Immune Estimation Resource (TIMER) database, which is based on data from TCGA (17). The TCGA-KIRC cohort comprised a total of 539 tumor and 72 adjacent normal tissue samples from ccRCC patients, and all the clinical information. The PANK1 mRNA expression levels in the tumor tissues and normal tissues from paired and unpaired patients in TCGA database were compared. Additionally, 2 Gene Expression Omnibus (GEO) data sets (https://www.ncbi.nlm.nih.gov/geo/) were used to analyze PANK1 expression patterns.
Correlation analysis of PANK1 expression and clinical characteristics
Correlation analyses of PANK1 mRNA expression and clinicopathological characteristics, including tumor (T) stage, node (N) stage, metastasis (M) status, pathological stage, histological grade, sex, and age, were performed in TCGA-KIRC cohort with R software using the “ggplot2” package. Meanwhile, the confounding effects of clinical factors were adjusted to ascertain the independent prognostic role of PANK1. Additionally, the expression levels of PANK1 in patients with different clinical stages from TCGA-KIRC cohort were calculated using the University of Alabama Cancer Database (UALCAN) website.
Overall and disease-specific survival analyses
The Gene Expression Profiling Interactive Analysis (GEPIA) website (available at http://gepia.cancer-pku.cn/) is a scientific tool to analyze clinical data from TCGA database and tissue-specific expression patterns (18). The online Kaplan-Meier plotter (available at http://kmplot.com/analysis/) can assess the role of individual genes on the survival prognosis of patients with cancer. The prognostic value of PANK1 expression in ccRCC patients was assessed using data from TCGA-KIRC cohort. OS, disease-specific survival (DSS), and progression-free survival (PFS) were determined by classifying the patients into 2 categories according to their median PANK1 expression level (high versus low). Log-rank P values and hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated. A ROC curve of diagnosis, a time-dependent ROC curve, and a nomogram model for prognosis analysis were created using the “pROC”, “timeROC”, and “survival” packages in R, respectively. The threshold value of area under curve (AUC) for a good prognosis predicting model is 0.75 according to previous study (19). All the clinical data used in this section were obtained from TCGA database.
Profiling of genes co-expressed with PANK1 and KEGG/GO analyses
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) analyses were performed using the co-expressed genes with the “clusterProfiler” package in R to explore the possible functions and signaling pathways affected by PANK1. The co-expression genes were screened using “limma” in R (20). The GO analysis included the biological process (BP), cellular composition (CC), and molecular function (MF); a two-sided P value <0.05 was considered statistically significant (21). A functional protein association network of PANK1 in Homo sapiens was generated by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) to display the protein-protein interaction networks (22).
Methylation analysis of PANK1 in ccRCC
An analysis of PANK1 promoter methylation status and clinical characteristics, including sample type, individual cancer stage, histological grade, sex, and age, was performed in the UALCAN database. Additionally, the online MethSurv tool was used to analyze the role of different methylation sites of PANK1 promoter with the survival data in TCGA cohort of ccRCC using the deoxyribonucleic acid (DNA) methylation data (23). Specifically, patients with ccRCC were divided into high- and low-risk groups according to the median methylation value of PANK1. The Xena platform was also used to analyze the correlation between the PANK1 expression and the methylation levels of certain methylation sites (24).
Correlation analysis of PANK1 and immune-infiltrating cells
The TIMER database was employed to analyze the levels and clinical significance of tumor-infiltrating immune cells (25), including cluster of differentiation (CD) 8+ T cells, CD4+ T cells, B cells, macrophages, neutrophils, and dendritic cells (DCs), and the relationship between these cell types and PANK1 expression was determined. The CIBERSORT (https://cibersort.stanford.edu/) (26) database was used to further quantify the infiltrating proportion of immune cells in ccRCC samples with high and low PANK1 expression. The “ggplot2,” “tidyverse,” and “reshape2” packages in R were used for the data analyses and graph plotting.
Cell culture, transfection, and cell viability assays
The ccRCC cell lines 786-O and CaKi-2 cells were obtained from the National Collection of Authenticated Cell Cultures (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium containing fetal bovine serum at a final concentration of 10% in a humidified atmosphere of 5% carbon dioxide. Small-interfering RNAs (siRNAs) against PANK1 and control siRNAs (scrambled siRNAs) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used in accordance with the manufacturer’s instructions. After transfection, Cell Counting Kit-8 (CCK-8) assays were performed as previously described (27).
Mitochondrial function measurements
The oxygen consumption rate (OCR) was analyzed using a 2-channel titration injection respirometer (Oxygraph-2k, Oroboros Instruments, Innsbruck, Austria) equipped with 2 chambers, as previously described (28). Additionally, mitoSOX Red (ThermoFisher) was used to determine mitochondrial-specific reactive oxygen species (ROS) production. To quantify the intensity of MitoSOX fluorescence, the plates were measured using a Fluoroskan Ascent Fluorometer (ThermoFisher, Helsinki, Finland).
Statistical analysis
SPSS (version 20.0) and R project (version 4.0.4) software were used for the statistical analyses. The clinical data are presented as the mean ± standard deviation (SD), while the cell experimental data are presented as the mean ± standard error mean (SEM). Independent samples t-tests and paired samples t-tests were performed to analyze differences in PANK1 mRNA levels between the tumor tissues and the adjacent normal tissues of ccRCC patients from the TCGA and GEO databases. The chi-squared test was used to analyze the associations between PANK1 expression levels and the clinical characteristic variables using TCGA data. Univariate and multivariate analyses were performed using the Cox proportional hazards regression model. For the details of multivariate Cox regression analysis, all the candidate clinical factors available in TCGA cohort were analyzed using the univariate cox analysis. The clinical factors with significant significance were included in the multivariate analyses with adjustments of confounding factors. The factors entered the multivariate Cox analysis model were considered as the outcome variables. OS, DSS, and PFS analyses were performed using Kaplan-Meier plots, and the differences were compared using the log-rank test. Gene expression associations were determined by a Spearman correlation analysis. Differences were considered statistically significant when the two-sided P value was <0.05.
Results
PANK1 expression is more decreased in ccRCC tissues than normal tissues
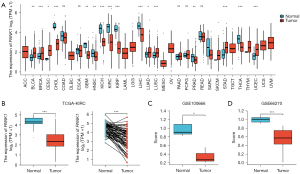
To further evaluate the PANK1 expression profiles in human tumors, we analyzed the TIMER database. PANK1 expression was more decreased in cholangiocarcinoma, colon adenocarcinoma, glioblastoma, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, prostate adenocarcinoma, and thyroid carcinoma tissues than normal tissues (see Figure 1A). Thus, we focused on the role of PANK1 in renal cancers. More specifically, in TCGA-KIRC cohort, which comprised a total of 539 tumor samples and 72 adjacent normal samples, the expression of PANK1 was decreased in the KIRC tissues in both the unpaired and paired data (P<0.001; see Figure 1B). Further, the expression patterns of PANK1 were verified using GEO data sets GSE100666 (N=3 in each group) and GSE66270 (N=14 in each group), and the results revealed that PANK1 expression was decreased in the tumor tissues than the normal control tissues (see Figure 1C,1D).
PANK1 expression is correlated with the clinical prognosis of ccRCC patients
According to the TCGA-KIRC data, patients with ccRCC in TCGA-KIRC cohort were divided into high (N=270) and low PANK1 (N=269) groups according to both our analysis and previous preprint (29). It showed that PANK1 expression levels were correlated with the key clinical characteristics. Next, the expression patterns of PANK1 at different clinical stages were analyzed in TCGA-KIRC cohort. Our results showed that PANK1 expression decreased as the T stage increased, and that there were significant differences between patients with stages T1 and T3 (P<0.001), and stages T1 and T4 (P<0.05; see Figure 2A). Additionally, N1-stage patients had lower PANK1 expression levels than N0-stage patients (P<0.05), and M1-stage patients also had lower PANK1 expression levels than M0-stage patients (P<0.01) (see Figure 2B,2C). Regarding pathological stage and histological grade, patients with a higher stage or grade had decreased PANK1 levels compared to those with a lower stage or grade (see Figure 2D,2E). Male patients had decreased PANK1 levels, but no significant differences were observed between older (>60 years) and younger (<60 years) patients (see Figure 2F,2G). Further, dead patients had decreased PANK1 levels compared to alive patients according to the OS, DSS, and progression-free interval (PFI) outcomes (see Figure 2H-2J). Univariate Cox analysis indicated that key clinical characteristics exhibited a significant correlation with clinical prognosis according to previous study (30). Patients with low PANK1 expression increased mortality compared with high PANK1 patients [hazards ratio (HR) =2.702, 95% confidence interval 1.957 to 3.731]; however, only M stage, histological stage, age, and PANK1 expression level were included in the multivariate analysis after adjusting for the confounding effects of clinical factors (see Table 1). These results indicate that PANK1 expression plays an important role in modulating clinical prognosis.
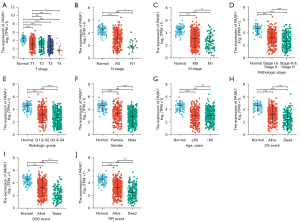
Table 1
| Characteristics | Total (N) | Multivariate analysis | |
|---|---|---|---|
| HR (95% CI) | P value | ||
| T stage | 539 | ||
| T1 & T2 | 349 | Reference | |
| T3 & T4 | 190 | 1.451 (0.634–3.323) | 0.378 |
| N stage | 257 | ||
| N0 | 241 | Reference | |
| N1 | 16 | 1.312 (0.651–2.646) | 0.448 |
| M stage | 506 | ||
| M0 | 428 | Reference | |
| M1 | 78 | 2.652 (1.567–4.486) | <0.001 |
| Pathologic stage | 536 | ||
| Stage I & II | 331 | Reference | |
| Stage III & IV | 205 | 1.212 (0.479–3.062) | 0.685 |
| Histologic grade | 531 | ||
| G1 & G2 | 249 | Reference | |
| G3 & G4 | 282 | 1.684 (1.020–2.778) | 0.041 |
| Age, years | 539 | ||
| ≤60 | 269 | Reference | |
| >60 | 270 | 1.665 (1.086–2.554) | 0.019 |
| PANK1 | 539 | ||
| High | 270 | Reference | |
| Low | 269 | 2.482 (1.546–3.986) | <0.001 |
CcRCC, clear cell renal cell carcinoma; HR, hazard ratio; CI, confidence interval; PANK1, pantothenate kinase-1.
PANK1 is an independent protective prognostic factor in ccRCC patients
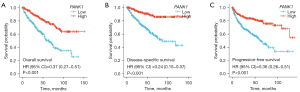
The prognostic value of PANK1 expression in patients with ccRCC was assessed by Kaplan-Meier plots and Log-rank analyses. Lower PANK1 levels were associated with significantly shortened OS (HR =0.37, P<0.001; see Figure 3A), DSS (HR =0.24, P<0.001; see Figure 3B), and PFS (HR =0.36, P<0.001; see Figure 3C) in TCGA-KIRC cohort. Thus, our results suggested that PANK1 is a protective prognostic factor and an independent prognostic marker for ccRCC.
Value of PANK1 expression in diagnosis and prognosis prediction
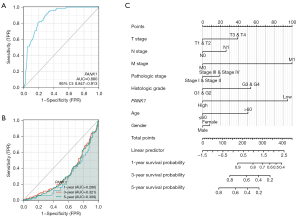
The ROC curve showed that PANK1 expression could be used to accurately differentiate between tumor and normal tissues (AUC =0.880) (see Figure 4A). Additionally, a time-dependent survival ROC curve of PANK1 was created to predict the 1-, 3-, and 5-year survival rates of patients. However, all these AUC values were lower than 0.6, which suggests that PANK1 expression cannot be used for clinical prediction (see Figure 4B). By integrating the clinicopathological factors (including age, sex, T stage, N stage, M stage, and histological and pathological stages) and PANK1 expression, a nomogram model was built to predict the survival probabilities at the 1-, 3-, and 5-year for patients in clinical practice. The nomogram model indicated that in addition to M stage, histologic stage, and age, PANK1 expression is a dominant factor in predicting the survival probability. (see Figure 4C).
Associations between PANK1 methylation status and clinical features in patients with ccRCC
First, we analyzed the promoter methylation levels of PANK1 and clinical features, and found that ccRCC tumors exhibited increased methylation levels compared to normal tissues (see Figure 5A). Additionally, pathological stage 1 had aberrant methylation levels compared to stage 3 or stage 4 (P<0.05) disease (see Figure 5B), while histological grade 4 had discrepant methylation levels compared to grade 2 (P<0.01) or stage 3 (P<0.05) disease (see Figure 5C). No significant differences in methylation levels were observed between N0 and N1, age, and sex (see Figure 5D-5F). Further, the DNA methylation levels of PANK1 were assessed according to the prognostic value of each single Cytosine-phosphate-Guanine (CpG) using the MethSurv tool. The results of MethSurv suggested a total of 14 methylation CpG sites, among which cg17051995 and cg26739829 had the highest DNA methylation levels (see Figure 5G). The methylation levels of 9 CpG sites (i.e., cg06326839, cg09161592, cg09747456, cg12226046, cg17051995, cg18222926, cg19519331, cg20349567, and cg26739829) were found to be correlated with clinical prognosis (P<0.05; see Table 2).
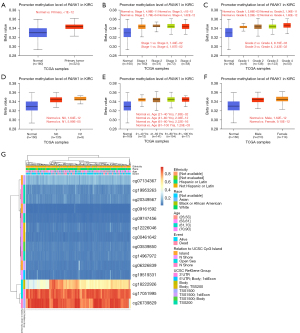
Table 2
| Gene symbol | CpG name | HR (95% CI) | LR test P value | UCSC ref gene group | Relation to UCSC CpG Island |
|---|---|---|---|---|---|
| PANK1 | cg00461642 | 0.643 (0.401, 1.032) | 0.067 | Body | N_Shore |
| cg03539850 | 1.396 (0.945, 2.064) | 0.094 | TSS1500 | N_Shore | |
| cg06326839 | 0.372 (0.232, 0.596) | 0.000039 | TSS1500 | Island | |
| cg07134367 | 1.287 (0.797, 2.077) | 0.3 | TSS1500 | Island | |
| cg09161592 | 0.485 (0.308, 0.764) | 0.0018 | TSS1500 | Island | |
| cg09747456 | 2.56 (1.727, 3.794) | 0.0000028 | TSS200 | S_Shore | |
| cg12226046 | 1.791 (1.078, 2.977) | 0.025 | Body | N_Shore | |
| cg14967972 | 1.263 (0.86, 1.856) | 0.23 | TSS200 | S_Shore | |
| cg17051995 | 0.485 (0.276, 0.853) | 0.012 | 3’UTR | Open_Sea | |
| cg18222926 | 2.186 (1.46, 3.275) | 0.00015 | Body | Open_Sea | |
| cg19519331 | 1.752 (1.181, 2.598) | 0.0053 | 5’UTR | N_Shore | |
| cg19953263 | 0.902 (0.575, 1.416) | 0.66 | TSS200 | S_Shore | |
| cg20349567 | 2.113 (1.418, 3.148) | 0.00024 | TSS200 | S_Shore | |
| cg26739829 | 1.862 (1.12, 3.094) | 0.016 | TSS1500 | S_Shore |
CpG, Cytosine-phosphate-Guanine; PANK1, pantothenate kinase-1; HR, hazard ratio; CI, confidence interval; LR, likelihood-ratio; UCSC, University of California Santa Cruz; TSS, transcription initiation site; UTR, untranslated region.
GO and KEGG pathway analyses of PANK1
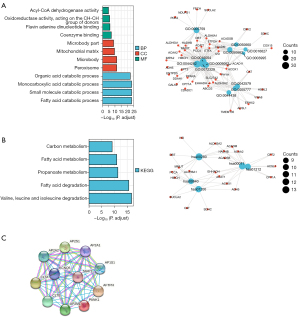
The top 100 most positively correlated genes with PANK1 in the GO and KEGG enrichment analyses were determined by the “clusterProfile” package in R. The GO analysis showed that most of the genes were associated with coenzyme binding, the mitochondrial matrix, peroxisome, and fatty acid catabolism (see Figure 6A). The KEGG data suggested that fatty acid metabolism may be related to the carcinogenic mechanism of PANK1 (see Figure 6B). In this study, PANK1-binding proteins were investigated using the STRING network, and the results showed that PANK1 interacts with ion channels, including the epidermal growth factor receptor (EGFR) pathway-, energy homeostasis-, and endocytosis-related proteins (see Figure 6C).
PANK1 is significantly correlated with tumor-infiltrating immune cells in ccRCC
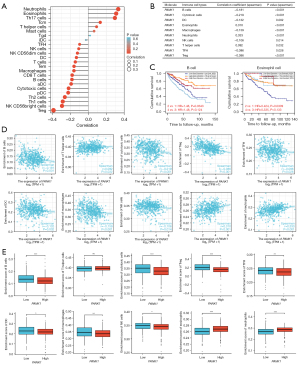
We further examined whether PANK1 expression was associated with the levels of immune cell infiltration in ccRCC. The single-sample gene set enrichment analysis tool from R and the Spearman’s R value were used to investigate the potential associations between the PANK1 expression and infiltration levels of 24 immune cell types. PANK1 expression was found to be significantly correlated with B cells, cytotoxic cells, DCs, eosinophils, macrophages, neutrophils, natural killer (NK) cells, T helper cells, Th1 cells, follicular helper T (TFH) cells, and Treg cells (see Figure 7A,7B). We also found that high levels of B cells and low levels of eosinophils were associated with the poor prognosis of ccRCC patients (P<0.05; see Figure 7C). Further research showed that PANK1 expression was positively correlated with the infiltration levels of T helper cells (r=0.092, P=0.032), eosinophils (r=0.310, P<0.001), and neutrophils (r=0.333, P<0.001). Conversely, PANK1 expression was negatively correlated with B cells (r=−0.181, P<0.001), cytotoxic cells (r=–0.219, P<0.0010.026), Tregs (r=−0.386, P<0.001), TFH cells (r=−0.096, P=0.026), DCs (r=−0.132, P=0.002), macrophages (r=−0.159, P<0.001), and NK cells (r=−0.106, P=0.014; see Figure 7D). Next, the relationship between PANK1 expression and immune infiltration was investigated. Somewhat surprisingly, we found significant differences in the levels of infiltrating immune cells, including B cells, cytotoxic cells, Tregs, TFH cells, DCs, macrophages, NK cells, eosinophils, and neutrophils (P<0.05) when PANK1 expression was categorized into high and low groups (see Figure 7E). These results indicated that PANK1 participates in immune cell infiltration in ccRCC.
Effects of PANK1 on the biological features of ccRCC cells
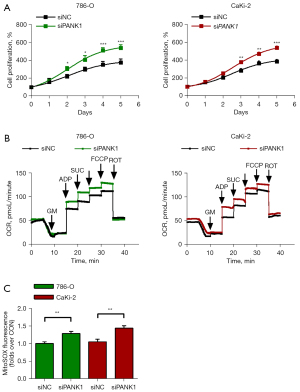
To analyze the effects of PANK1 on the biological features of ccRCC, 2 cell lines (i.e., 786-O and CaKi-2) were used for further experiments. We found that the knockdown of PANK1 promoted cell proliferation in both the 786-O and CaKi-2 cells (see Figure 8A). Additionally, the OCR values of these 2 cell lines were measured, and the results revealed that PANK1-specific siRNA (siPANK1) increased the OCR and mitochondrial-specific ROS production (see Figure 8B,8C). These results indicate that PANK1 regulates ccRCC pathogenesis by modulating the global metabolism of tumor cells.
Discussion
CcRCC is a type of severe malignant tumor and has increased in incidence in recent years (1). Great advances in the understanding the molecular mechanisms of ccRCC pathogenesis and progression have led to unprecedented progress in the diagnosis of and therapeutic strategies for ccRCC (31). Developments have been made in the treatment of ccRCC, including surgical resection, molecular-targeted drugs (vascular endothelial growth factor, platelet-derived growth factor, EGFR, and mTOR), and immunotherapies targeting the programmed death-ligand 1/programmed death-1 pathway in advanced ccRCC (32). However, the therapeutic effects are still unsatisfactory, and as ccRCC is a heterogeneous disease, more research needs to be conducted to identify novel targets to enable the better diagnosis, therapy, and prognosis of ccRCC.
In this study, we mainly focused on PANK1, an enzyme critical in the regulation of global metabolism, whose role in ccRCC and other tumors is still unknown. Using TCGA and GEO databases, we confirmed that PANK1 expression is more decreased in ccRCC tissues than normal renal tissues. Promoter methylation status may contribute to this expression discrepancy. We also found that PANK1 expression was significantly correlated with TNM stage, pathological stage, histological grade, and survival status. Additionally, its expression was correlated with the immune filtration status of several crucial immune cell types. Thus, PANK1 expression was shown to have a good ability to distinguish between tumor tissues and normal tissues.
A pan-cancer analysis showed that PANK1 was downregulated in all types of renal cancers. Further research revealed that low PANK1 levels were associated with reduced OS, DSS, and PFS in ccRCC patients. The logistic regression demonstrated that PANK1 expression level was correlated with the clinicopathological characteristics of ccRCC. Additionally, the univariate and multivariate Cox analyses indicated that PANK1 was an independent positive factor for predicting clinical prognosis. These data and the results of the ROC analysis suggested that PANK1 may be a promising prognostic biomarker for ccRCC patients. Further, while gene mutations are closely related to tumor pathogenesis and are often associated with a bad prognosis, the percentage of PANK1 genetic alterations in ccRCC was only approximately 0.5% (cBioPortal, www.cbioportal.org; data not provided). Additionally, changes in DNA methylation status are common epigenetic mechanisms in almost all forms of cancer. The relationship between the DNA methylation levels of PANK1 and the prognosis of ccRCC patients was investigated, and we found that promoter methylation was associated with the pathological stage and histological grade. The hypermethylation of 9 CpG sites was correlated with a worse OS, and contributed to decreased PANK1 expression levels in ccRCC patients, especially among those with a poor prognosis.
The tumor microenvironment (TME) is composed of various types of immune cells in tumor and plays an important role in tumor progression, metastasis, and treatment response (33). The composition of tumor-infiltrating immune cells strongly affects the TME, tumor behavior, and clinical prognosis. As PANK1 is a critical gene in cell metabolism regulation, we hypothesized that PANK1 affects the TME by modulating the proportions and infiltration of specific immune cell types. A recent study showed that PANK1 is associated with immune cell infiltration (16). In the present study, we systematically analyzed the effects of PANK1 on immune cell infiltration, and found that Tregs, cytotoxic cells, B cells, macrophages, DCs, NK cells, and TFH cells were negatively correlated with PANK1 expression, while neutrophils, eosinophils, and T helper cells were positively correlated with PANK1 expression. More importantly, high PANK1-expressing or low PANK1-expressing tumors had aberrant patterns of immune cell infiltration rates. Certainly, the regulation of the TME is highly of complex, and the proportions of immune cell types in the TME may also affect tumor cell survival. Future studies need to be conducted to further explore the relationship between PANK1 and these different cell types.
To determine the BPs in which PANK1 is involved, a functional analysis was conducted using GO/KEGG and STRING. Consistent with the function of PANK1 as a critical gene in global metabolism regulation, we found that most PANK1-related genes were associated with coenzyme binding, the mitochondrial matrix, peroxisome, and fatty acid metabolism, and may also be related to carcinogenesis. A previous study indicated that PANK1 is a crucial rate-limiting enzyme in the de-novo synthesis of CoA, which controls the ratio and rate of CoA to acetyl-CoA. The systemic knockout of PANK1 and PANK2 in neurons results in a decrease in the content of short-chain acyl-CoA (14). In Lep-knockout mice, the knockout of Pank1 alleviated the hyperglycemia and hyperinsulinemia phenotypes (34). Leonardi et al. demonstrated the importance of PANK1 during fasting, which supports the metabolic transition from glucose use and fatty acid synthesis to gluconeogenesis and fatty acid oxidation, and suggests that PANK1 could be a therapeutic target for metabolic disorders (35).
Additionally, as PANK1 is considered an important downstream target of p53 transcription (13,15,36), its role in carcinogenesis has been investigated in recent years. PANK genes have been shown to have prognostic significance in acute myeloid leukemia using TCGA database (37). A recent study revealed that the overexpression of PANK1 inhibits the proliferation, growth, invasion, and tumorigenicity of hepatocellular carcinoma cells by inhibiting Wnt/β-catenin signaling (11). However, the role of PANK1 in other tumors has not yet been investigated. In our study, we found that PANK1 was significantly associated with clinical prognosis, and PANK1 knockdown promoted cell proliferation in ccRCC.
The role of energy metabolism has been shown to play dominant roles in both cancer pathogenesis and progression. Findings that metabolic demands change frequently during tumor initiation, progression, and metastasis have challenged our understanding of tumor biology and the development of novel therapeutics. Thus, investigating the role of TME cells in the remodeling of cancer cell energy metabolism has become an increasingly important area of research (38). Additionally, metabolic changes have been shown to occur in ccRCC. Notably, Wu et al. reported that redox-related genes were associated with immune infiltration and could accurately predict ccRCC prognosis (39). Xing et al. identified 10 glycolysis-related genes that predicted OS for ccRCC patients (40). In the present study, we found that PANK1 knockdown regulated mitochondrial bioenergetics and redox status, which is a critical mechanism underlying PANK1-mediated effects on ccRCC. Tumor-associated immune cells and immune checkpoint inhibition play important roles in the carcinogenesis and treatment of ccRCC (41). PANK1 has been shown to be correlated with the infiltration status of several types of immune cells. Thus, PANK1 may be an important link between metabolism and immunity during cancer pathogenesis and treatment.
This study was the first to systematically explore the relationship between PANK1 expression and ccRCC; however, it had some limitations. First, most of the clinical and expression data analyzed by bioinformatics were downloaded from TCGA or GEO databases, and our findings require further validation by systematical experimental investigations. Second, the immune infiltration status regulated by PANK1 requires further experimental exploration. Third, longer follow-up durations and larger cohorts of patients are needed to validate that PANK1 is an accurate prognosis predictor.
Conclusions
In this study, we verified for the first time the value of PANK1 in the prognosis prediction of ccRCC. The DNA methylation of PANK1 is related to the prognosis of ccRCC. PANK1 not only participates in the occurrence and progression of ccRCC but also its immune regulation. Additionally, PANK1 associates with mitochondrial metabolism and may be an important link between tumor metabolism and immunity. Thus, PANK1 could be a potential prognostic biomarker and a promising therapeutic target for ccRCC. Further mechanistic studies need to be conducted to validate our findings and promote its clinical application.
Acknowledgments
We would like to thank H. Nikki March, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
Funding: This study was funded by the Specialized Scientific Program of the Innovation Platform for Academicians of Hainan Province (YSPTZX202026), the Hainan Provincial Natural Science Foundation of China (821QN383), the Specialized Scientific Research Project of Military Health Care (21BJZ37), the NSFC General Project (81570679), Beijing NOVA program (Z161100004916141), and the Innovation Platform for Academicians of Hainan Province (Academician Chen Xiangmei of Hainan Province Kidney Diseases Team Innovation Center).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1488/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1488/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1488/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Choueiri TK, Halabi S, Sanford BL, et al. Cabozantinib Versus Sunitinib As Initial Targeted Therapy for Patients With Metastatic Renal Cell Carcinoma of Poor or Intermediate Risk: The Alliance A031203 CABOSUN Trial. J Clin Oncol 2017;35:591-7. [Crossref] [PubMed]
- Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380:1103-15. [Crossref] [PubMed]
- Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380:1116-27. [Crossref] [PubMed]
- Wu Y, Zhang X, Wei X, et al. A Mitochondrial Dysfunction and Oxidative Stress Pathway-Based Prognostic Signature for Clear Cell Renal Cell Carcinoma. Oxid Med Cell Longev 2021;2021:9939331. [Crossref] [PubMed]
- Yao Z, Zheng Z, Zheng X, et al. Comprehensive Characterization of Metabolism-Associated Subtypes of Renal Cell Carcinoma to Aid Clinical Therapy. Oxid Med Cell Longev 2022;2022:9039732. [Crossref] [PubMed]
- Farber NJ, Kim CJ, Modi PK, et al. Renal cell carcinoma: the search for a reliable biomarker. Transl Cancer Res 2017;6:620-32. [Crossref] [PubMed]
- Petitprez F, Ayadi M, de Reyniès A, et al. Review of Prognostic Expression Markers for Clear Cell Renal Cell Carcinoma. Front Oncol 2021;11:643065. [Crossref] [PubMed]
- Qi X, Li Q, Che X, et al. The Uniqueness of Clear Cell Renal Cell Carcinoma: Summary of the Process and Abnormality of Glucose Metabolism and Lipid Metabolism in ccRCC. Front Oncol 2021;11:727778. [Crossref] [PubMed]
- Chen C, Zhao W, Lu X, et al. AUP1 regulates lipid metabolism and induces lipid accumulation to accelerate the progression of renal clear cell carcinoma. Cancer Sci 2022; [Epub ahead of print]. [Crossref] [PubMed]
- Zi Y, Gao J, Wang C, et al. Pantothenate Kinase 1 Inhibits the Progression of Hepatocellular Carcinoma by Negatively Regulating Wnt/β-catenin Signaling. Int J Biol Sci 2022;18:1539-54. [Crossref] [PubMed]
- Czumaj A, Szrok-Jurga S, Hebanowska A, et al. The Pathophysiological Role of CoA. Int J Mol Sci 2020;21:9057. [Crossref] [PubMed]
- Wang SJ, Yu G, Jiang L, et al. p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle 2013;12:753-61. [Crossref] [PubMed]
- Subramanian C, Yao J, Frank MW, et al. A pantothenate kinase-deficient mouse model reveals a gene expression program associated with brain coenzyme a reduction. Biochim Biophys Acta Mol Basis Dis 2020;1866:165663. [Crossref] [PubMed]
- Böhlig L, Friedrich M, Engeland K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res 2011;39:440-53. [Crossref] [PubMed]
- Zhang Y, Tang M, Guo Q, et al. The value of erlotinib related target molecules in kidney renal cell carcinoma via bioinformatics analysis. Gene 2022;816:146173. [Crossref] [PubMed]
- Zhao K, Ma Z, Zhang W. Comprehensive Analysis to Identify SPP1 as a Prognostic Biomarker in Cervical Cancer. Front Genet 2022;12:732822. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chen J, Wang H, Zhou L, et al. A combination of circulating tumor cells and CA199 improves the diagnosis of pancreatic cancer. J Clin Lab Anal 2022;36:e24341. [Crossref] [PubMed]
- Shang BB, Chen J, Wang ZG, et al. Significant correlation between HSPA4 and prognosis and immune regulation in hepatocellular carcinoma. PeerJ 2021;9:e12315. [Crossref] [PubMed]
- Zhou Q, Hou Z, Zuo S, et al. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci 2019;110:1194-207. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Wang B, Ni Z, Dai X, et al. The Bcl-2/xL inhibitor ABT-263 increases the stability of Mcl-1 mRNA and protein in hepatocellular carcinoma cells. Mol Cancer 2014;13:98. [Crossref] [PubMed]
- Wang B, Xiong S, Lin S, et al. Enhanced Mitochondrial Transient Receptor Potential Channel, Canonical Type 3-Mediated Calcium Handling in the Vasculature From Hypertensive Rats. J Am Heart Assoc 2017;6:005812. [Crossref] [PubMed]
- Ma B, Wang P. PANK1 is a Prognostic Biomarker Associated with Immune Infiltration of Clear Cell Renal Carcinoma. Research Square. [Preprint.] Oct 13, 2021 [accessed 2022 June 6]. Available online: https://doi.org/
10.21203/rs.3.rs-957747/v1 . - Cui Y, Zhou Z, Chai Y, et al. Upregulated GSDMB in Clear Cell Renal Cell Carcinoma Is Associated with Immune Infiltrates and Poor Prognosis. J Immunol Res 2021;2021:7753553. [Crossref] [PubMed]
- Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801-6. [Crossref] [PubMed]
- Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8:793-800. [Crossref] [PubMed]
- Kim MC, Jin Z, Kolb R, et al. Updates on Immunotherapy and Immune Landscape in Renal Clear Cell Carcinoma. Cancers (Basel) 2021;13:5856. [Crossref] [PubMed]
- Leonardi R, Rock CO, Jackowski S. Pank1 deletion in leptin-deficient mice reduces hyperglycaemia and hyperinsulinaemia and modifies global metabolism without affecting insulin resistance. Diabetologia 2014;57:1466-75. [Crossref] [PubMed]
- Leonardi R, Rehg JE, Rock CO, et al. Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One 2010;5:e11107. [Crossref] [PubMed]
- Yang L, Zhang B, Wang X, et al. P53/PANK1/miR-107 signalling pathway spans the gap between metabolic reprogramming and insulin resistance induced by high-fat diet. J Cell Mol Med 2020;24:3611-24. [Crossref] [PubMed]
- Liu Y, Cheng Z, Li Q, et al. Prognostic significance of the PANK family expression in acute myeloid leukemia. Ann Transl Med 2019;7:261. [Crossref] [PubMed]
- Herst PM, Carson GM, Eccles DA, et al. Bioenergetic and Metabolic Adaptation in Tumor Progression and Metastasis. Front Oncol 2022;12:857686. [Crossref] [PubMed]
- Wu Y, Wei X, Feng H, et al. Integrated Analysis to Identify a Redox-Related Prognostic Signature for Clear Cell Renal Cell Carcinoma. Oxid Med Cell Longev 2021;2021:6648093. [Crossref] [PubMed]
- Xing Q, Zeng T, Liu S, et al. A novel 10 glycolysis-related genes signature could predict overall survival for clear cell renal cell carcinoma. BMC Cancer 2021;21:381. [Crossref] [PubMed]
- Zarrabi K, Walzer E, Zibelman M. Immune Checkpoint Inhibition in Advanced Non-Clear Cell Renal Cell Carcinoma: Leveraging Success from Clear Cell Histology into New Opportunities. Cancers (Basel) 2021;13:3652. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


