
Development and validation of an online prognostic nomogram for osteosarcoma after surgery: a retrospective study based on the SEER database and external validation with single-center data
Introduction
Osteosarcoma accounts for about 35% of the primary malignancy of bone, which remains the most common type of bone cancer (1,2). The multidisciplinary surgical center team approach has been well accepted for the treatment of osteosarcoma (3). With the development of high-intensity combination chemotherapy and surgical techniques since the 1970s, the 5-year survival rate for patients has increased from 20% to approximately 60% (4,5). However, due to their heterogeneous therapeutic response, 30–40% of these patients experience relapse and achieve a poor prognosis with a long-term survival rate of less than 20% (6). Thus, orthopedists and oncologists need to accurately estimate prognoses, which can ensure the implementation of appropriate treatment.
The Enneking staging system and the American Joint Committee on Cancer (AJCC) TNM staging system are generally used to roughly assess the clinical risks of osteosarcoma; nevertheless, they ignore the wide clinical heterogeneity of individuals and cannot accurately estimate prognosis. Previous studies have focused on various prognostic factors, such as age (7,8), tumor size and site (9), extent of disease (9,10), tumor grade (10), post-chemotherapeutic necrosis rate/Huvos classification (7), pathological fracture (11) and therapeutic methods (3,5); some recently reported techniques like radiomics have also revealed their prognostic significance with satisfactory accuracy in osteosarcoma (12). However, due to the low incidence (4 to 6 cases per million worldwide) of osteosarcoma (1), existing studies have been often limited to small sample sizes, which may lead to inevitable overfitting; and survival prediction models for osteosarcoma are relatively rare (13-15). Thus, a large cohort of patients and an integration of prognostic factors can be helpful in establishing a statistical prediction model.
The Surveillance, Epidemiology, and End Results (SEER) database is a large population-based dataset, which contains cancer incidence and survival information from 18 cancer registries covering 28% of the US population (16); therefore, the SEER database is a useful tool for the analysis of rare cancers.
Nomograms can visually integrate various factors to predict a specific endpoint; thus, recent studies have utilized this tool to achieve individual prognostic assessment and to quantify risks in various cancers (17,18).
In this study, using a large sample from the SEER database to explore the clinicopathological factors that predict postoperative survival for osteosarcoma. We developed a nomogram that facilitates individual prediction, and conducted external validation of single-center data in an Asian/Chinese population for clinical application. To our knowledge, no prognostic nomogram in osteosarcoma with independent validation has been reported yet in an Asian/Chinese population and no online version has been established. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2756/rc).
Methods
Data sources
Data for the training set and the internal validation set were retrospectively obtained from the SEER database (SEER Research Plus Data, 18 Registries, Nov 2020 Sub. 2000–2018) using SEER*Stat software 8.3.9.1 (National Cancer Institute, Bethesda, USA) (16). The access to individual data in this study was approved by the National Cancer Institute (accession number: 16781-Nov2020). The SEER database is publicly available and has removed personal information identification, and thus complies with the ethics committee requirements for biomedical research.
Data for external validation were retrospectively collected from patients with osteosarcoma in our hospital (Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology), from 2013 to 2016.
Inclusion and exclusion criteria
Inclusion criteria were as follows: (I) year of diagnosis from 2000 to 2016, to ensure a long-term follow-up information for 5 years; (II) diagnosis of osteosarcoma, ICD-O-3 Hist/behav including 9180/3 osteosarcoma not otherwise specified (NOS), 9181/3 chondroblastic osteosarcoma, 9182/3 fibroblastic osteosarcoma, 9183/3 telangiectatic osteosarcoma, 9184/3 osteosarcoma in Paget disease of bone, 9185/3 small cell osteosarcoma, 9186/3 central osteosarcoma, 9187/3 intraosseous well-differentiated osteosarcoma, 9192/3 parosteal osteosarcoma, 9193/3 periosteal osteosarcoma, and 9194/3 high-grade surface osteosarcoma; (III) diagnostic confirmation is macroscopically confirmed; (IV) pathological diagnosis with the first malignant primary indicator; and (V) resection of the primary tumor, site-specific surgery codes including 10–90 (not including 00 none, no surgery of primary site, or autopsy only; 99 unknown).
Individual patient data were excluded if (I) they lacked of general information and major information on clinicopathological characteristics, including age, sex, primary site, tumor size, and AJCC-T/N/M; (II) extra-skeletal site of primary tumor; (III) no specific cause of death; or (IV) survival time less than 1 month.
Data extraction and endpoint definition
Demographic and clinicopathological data, including age at diagnosis, sex, race, marital status at diagnosis, ICD-O-3 Hist/behave, primary site, laterality, tumor size, and tumor extension, histological grade, AJCC-stage group, AJCC-T, AJCC-N, AJCC-M, surgery, radiation recode, and chemotherapy recode, were obtained. The 6th AJCC TNM staging guidelines, which included AJCC-T, AJCC-N, AJCC-M, and grade, were used in our study (19). Marital status was characterized as single (never married), married, and divorced/widowed/separated. Primary site codes were classified into extremities (C40.0, C40.1, C40.2, C40.3, C40.8, and C40.9), pelvis/spine (C41.2 and C41.4), and skull/mandible/other (C41.0, C41.1, C41.3, C41.8, and C41.9) (15). Histological grade I, II, III, and IV represented well differentiated, moderately differentiated, poorly differentiated, and undifferentiated/anaplastic, respectively. Grades I and II were defined as low-grade osteosarcoma, while grades III and IV were converted to high-grade osteosarcoma (20,21). Based on surgery codes (https://seer.cancer.gov/manuals/2021/AppendixC/Surgery_Codes_Bones_2021.pdf), the types of surgery were classified into local or partial resection (codes 15, 19, 25, 26), radical resection with limb salvage (code 30), amputation (codes 40, 41, 42, 50), forequarter/hindquarter/hemipelvectomy (codes 51, 52, 53, 54), and surgery, NOS (code 90) (22,23).
Data from the external validation set were extracted from the medical record of our hospital. Data collection followed the same inclusion and exclusion criteria.
The endpoints were cancer-specific survival (CSS) and overall survival (OS). CSS was defined as the time from diagnosis to the occurrence of cancer-related deaths; OS was defined as the time from diagnosis until death from any cause or the last follow-up. Follow-up procedures were based on SEER database files; in our institution, patients are recommended for routine followed-up (every 3 months during the first 2 years, every 6 months for up to 5 years, and then annually thereafter). In August 2021, the last follow-up assessment with telephone investigations of patients in our institution was supplemented.
Statistical analysis
Data obtained from the SEER database were divided into a training set and an internal validation set according to the random number generation method (at a 1:1 ratio). The training set was used to determine independent prognostic factors and construct the nomograms. The internal and external validation sets were used to validate the nomograms. SPSS 25.0 software (IBM Corp., USA), and R 4.1.1 software (R core team, Austria) were used to process the data. A two-sided P≤0.05 was considered statistically significant. The chi-squared test was used to compare differences in categorical variables between groups; the Student’s t-test was used to compare averages for continuous variables. All variables with statistical significance in the univariate analysis were considered in a multivariate Cox proportional hazard model to identify independent predictors. The outcomes were presented as the hazard ratio (HR) with 95% confidence intervals (CIs).
The ‘rms’ and ‘survival’ packages in R software were used to establish the nomograms of independent predictors (17). The ‘DynNom’ package was used to build an online version of our nomogram. The discrimination and calibration of the nomograms were evaluated using the Harrell concordance index (C-index) and the calibration curves, for the three sets. The C-index was calculated using the coxph function in the ‘survival’ package; a higher C-index represented a more accurate prediction. The ‘rms’, ‘pec’ and ‘dplyr’ packages were used to generate the calibration curves. The patients in the training set, the internal validation set and the external validation set were divided into quartiles (n=1,057, 260 subjects per group for the first two sets; n=65, 16 subjects per group for the external validation set); 1000 bootstrap was selected to guide the resampling to quantify any overfitting and so compare the probabilities predicted by the nomogram with the actual survival results observed by Kaplan-Meier analysis (17). Decision curve analysis (DCA) was used to evaluate the clinical benefits of the nomogram using the ‘rms’ and ‘ggDCA’ packages. Compared to traditional metrics such as the area under the curve (AUC) that focuses solely on the predictive precision of a single statistic, DCA provides information on the clinical value of the model (24). In addition, comparison between our nomograms and the 6th AJCC TNM staging was conducted by calculating AUC using time-dependent receiver operating curve (tdROC), which was achieved through the ‘timeROC’ package (25). Details of the R code used to generate our nomogram were available in https://cdn.amegroups.cn/static/public/tcr-21-2756-1.pdf.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology [No. 2022 (0032)] and individual consent for this retrospective analysis was waived.
Results
Clinicopathological characteristics
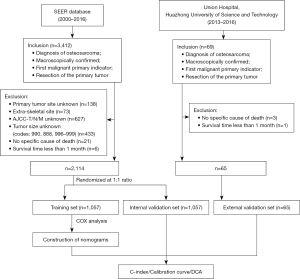
The flow diagram showing the inclusion and exclusion process is summarized in Figure 1. Using the stated inclusion criteria, data for 3,412 patients with osteosarcoma were obtained from the SEER database. Among these patients, 1,298 patients were excluded according to the exclusion criteria. The final number of patients included in the study was 2,114. According to the random number generation method (SPSS, fix value =2,000,000), 2,114 patients were divided into a training set (n=1,057) and an internal validation set (n=1,057) at a 1:1 ratio. In addition, data for 65 patients were obtained from our hospital as the external validation set.
The clinicopathological characteristics of the patients are shown in Table 1. No statistically significant differences were found in the baseline characteristics across the three datasets (all P>0.05 except race). Demographically, the mean age was 25.17±17.55, 24.61±17.70, and 26.96±14.06 years in the training, internal validation, and external validation sets, respectively. The training set comprised 55.6% males and 76.5% white patients, the internal validation set comprised 53.2% males and 74.5% white patients, while the external validation set comprised 58.5% males and entirely of Asians. Histologically, the type of osteosarcoma, NOS, represented 60.0%, 65.6%, and 66.2% in the training, internal validation, and external validation sets, respectively. Meanwhile, 83.3%, 81.2%, and 81.5% patients were located in the extremities in the training, internal validation, and external validation sets, respectively. Therapeutically, more than half of the patients (61.2%) underwent radical resection with salvage of the limb and most of the patients (85.0%) received chemotherapy in the training set, with a similar distribution in the two validation sets. In the entire cohort, the median follow-up time was 70.62±48.65 months. The 1-, 3- and 5-year CSS rates were 94.4% (95% CI: 93.4–95.4%), 76.7% (95% CI: 74.9–78.5%), and 69.4% (95% CI: 67.4–71.4%), respectively; the 1-, 3- and 5-year OS rates were 93.6% (95% CI: 92.6–94.6%), 75.5% (95% CI: 73.7–77.3%), and 67.7% (95% CI: 65.7–69.7%), respectively.
Table 1
| Characteristics | Training set (n=1,057) | Internal validation set (n=1,057) | P1 | External validation set (n=65) | P2 |
|---|---|---|---|---|---|
| Age (years) | 25.17±17.55 | 24.61±17.70 | 0.461 | 26.96±14.06 | 0.333 |
| Sex | 0.256 | 0.655 | |||
| Male | 588 (55.6) | 562 (53.2) | 38 (58.5) | ||
| Female | 469 (44.4) | 495 (46.8) | 27 (41.5) | ||
| Race | 0.627 | <0.001 | |||
| White | 809 (76.5) | 787 (74.5) | 0 (0.0) | ||
| Black | 151 (14.3) | 171 (16.2) | 0 (0.0) | ||
| Asian or Pacific Islander | 88 (8.3) | 88 (8.3) | 65 (100.0) | ||
| American Indian | 9 (0.9) | 11 (1.0) | 0 (0.0) | ||
| Marital status | 0.208 | 0.089 | |||
| Single | 793 (75.0) | 825 (78.1) | 42 (64.6) | ||
| Married | 220 (20.8) | 188 (17.8) | 21 (32.3) | ||
| Divorced/widowed/separated | 44 (4.2) | 44 (4.2) | 2 (3.1) | ||
| ICD-O-3/pathological subtype | 0.287 | 0.573 | |||
| 9180/3 osteosarcoma (NOS) | 634 (60.0) | 693 (65.6) | 43 (66.2) | ||
| 9181/3 chondroblastic osteosarcoma | 175 (16.6) | 147 (13.9) | 12 (18.5) | ||
| 9182/3 fibroblastic osteosarcoma | 62 (5.9) | 41 (3.9) | 6 (9.2) | ||
| 9183/3 telangiectatic osteosarcoma | 44 (4.2) | 37 (3.5) | 2 (3.1) | ||
| 9184/3 osteosarcoma in Paget disease of bone | 5 (0.5) | 4 (0.4) | 0 (0.0) | ||
| 9185/3 small cell osteosarcoma | 9 (0.9) | 12 (1.1) | 1 (1.5) | ||
| 9186/3 central osteosarcoma | 40 (3.8) | 46 (4.4) | 0 (0.0) | ||
| 9187/3 intraosseous well-differentiated osteosarcoma | 2 (0.2) | 2 (0.2) | 0 (0.0) | ||
| 9192/3 parosteal osteosarcoma | 62 (5.9) | 52 (4.9) | 1 (1.5) | ||
| 9193/3 periosteal osteosarcoma | 18 (1.7) | 16 (1.5) | 0 (0.0) | ||
| 9194/3 high-grade surface osteosarcoma | 6 (0.6) | 7 (0.7) | 0 (0.0) | ||
| Primary site | 0.198 | 0.445 | |||
| Extremities | 881 (83.3) | 858 (81.2) | 53 (81.5) | ||
| Pelvis/spine | 59 (5.6) | 55 (5.2) | 6 (9.2) | ||
| Skull/mandible and others | 117 (11.1) | 144 (13.6) | 6 (9.2) | ||
| Grade | 0.877 | 0.550 | |||
| Well/moderately differentiated; Grade I/II | 208 (19.7) | 205 (19.4) | 10 (15.4) | ||
| Poorly/undifferentiated/anaplastic; Grade III/IV | 746 (70.6) | 742 (70.2) | 50 (76.9) | ||
| Unknown | 103 (9.7) | 110 (10.4) | 5 (7.7) | ||
| Laterality | 0.295 | 0.120 | |||
| Not a paired site (axial) | 98 (9.3) | 117 (11.1) | 11 (16.9) | ||
| Left-origin of primary | 505 (47.8) | 475 (44.9) | 30 (46.2) | ||
| Right-origin of primary | 454 (43.0) | 464 (43.9) | 24 (36.9) | ||
| Bilateral, single primary | 0 (0.0) | 1 (0.1) | 0 (0.0) | ||
| AJCC-stage | 0.640 | 0.097 | |||
| I | 278 (26.3) | 281 (26.6) | 10 (15.4) | ||
| II | 600 (56.8) | 605 (57.2) | 38 (58.5) | ||
| III | 14 (1.3) | 17 (1.6) | 2 (3.1) | ||
| IV | 165 (15.6) | 154 (14.6) | 15 (23.1) | ||
| AJCC-T | 0.265 | 0.078 | |||
| T1 | 433 (41.0) | 470 (44.5) | 23 (35.4) | ||
| T2 | 594 (56.2) | 558 (52.8) | 37 (56.9) | ||
| T3 | 30 (2.8) | 29 (2.7) | 5 (7.7) | ||
| AJCC-N | 0.156 | 0.330 | |||
| N0 | 1041 (98.5) | 1032 (97.6) | 63 (96.9) | ||
| N1 | 16 (1.5) | 25 (2.4) | 2 (3.1) | ||
| AJCC-M | 0.417 | 0.252 | |||
| M0 | 901 (85.2) | 914 (86.5) | 52 (80.0) | ||
| M1 | 156 (14.8) | 143 (13.5) | 13 (20.0) | ||
| Surgery code | 0.692 | 0.284 | |||
| Local or partial resection | 149 (14.1) | 151 (14.3) | 8 (12.3) | ||
| Radical resection with limb salvage | 647 (61.2) | 672 (63.6) | 34 (52.3) | ||
| Amputation | 212 (20.1) | 186 (17.6) | 19 (29.2) | ||
| Forequarter/hindquarter/hemipelvectomy | 39 (3.7) | 39 (3.7) | 4 (6.2) | ||
| Surgery, NOS | 10 (0.9) | 9 (0.9) | 0 (0) | ||
| Radiation | 0.669 | 0.860 | |||
| None/unknown | 986 (93.3) | 981 (92.8) | 61 (93.8) | ||
| Yes | 71 (6.7) | 76 (7.2) | 4 (6.3) | ||
| Chemotherapy | 0.717 | 0.103 | |||
| No/unknown | 159 (15.0) | 165 (15.6) | 5 (7.7) | ||
| Yes | 898 (85.0) | 892 (84.4) | 60 (92.3) | ||
| Tumor size (mm) | 100.91±67.52 | 98.19±58.58 | 0.322 | 85.97±36.65 | 0.077 |
| Extension (mm) | 35.87±14.85 | 36.31±15.49 | 0.505 | 37.38±19.46 | 0.435 |
No. of patients (%) for categorical variable; mean ± standard deviation for continuous variables. P1 refers to the difference between training set and internal verification set; P2 refers to the difference between training set and external validation set. AJCC, the American Joint Committee on Cancer; NOS, not otherwise specified.
Univariate and multivariate analysis of prediction factors
Considering that AJCC-Stage is composed of grade and AJCC-T/N/M, these variables form a high degree of collinearity (Table S1) (26,27); thus the AJCC-Stage will not be included in univariate and multivariate analysis. Univariate analysis suggested that age, sex, pathological subtype (ICD-O-3 Hist/behave), primary site, grade, AJCC-T, AJCC-N, AJCC-M, tumor size, tumor extension, surgery type, radiation, and chemotherapy were significantly associated with CSS (all P≤0.05), whereas race, marital status, and laterality failed to reach statistical significance (P>0.05) (Table 2). After excluding non-significant variables, the remaining variables were used in the multivariate Cox proportional hazard model. Multivariate analysis revealed that only older age, osteosarcoma NOS, AJCC-N1, AJCC-M1, radiation treatment, larger tumor size, and longer tumor extension were independent prognostic factors of unfavorable CSS (all P≤0.05) (Table 2). Interestingly, patients who received radiotherapy or chemotherapy had worse CSS, which we believe may have introduced a bias because the SEER database was drawn from real-world data where more chemotherapy or radiotherapy generally associated with more severe disease severity. The relationship between radiotherapy/chemotherapy and clinicopathological characteristics is described in Table S2. Chemotherapy was significantly associated with several characteristics of poor prognosis, such as Grade III/IV, fewer AJCC-Stage I, more AJCC-Stage IV, fewer AJCC-T1, more AJCC-M1, larger tumor size, and longer tumor extension (all P<0.05). To further confirm the credibility of models, we repeated the Cox multivariate analysis excluding radiotherapy and chemotherapy, and obtained the same predictive factors (Tables S3,S4).
Table 2
| Characteristics | Univariate analyses | Multivariate analyses | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (years) | 1.015 (1.010–1.021) | <0.001* | 1.027 (1.021–1.034) | <0.001* | |
| Sex | 0.001* | 0.111 | |||
| Male | 1.000 | 1.000 | |||
| Female | 0.689 (0.552–0.859) | 0.830 (0.660–1.044) | |||
| Race | 0.069 | – | |||
| White | 1.000 | – | |||
| Black | 0.838 (0.601–1.169) | – | |||
| Asian or Pacific Islander | 1.314 (0.923–1.871) | – | |||
| American Indian | 2.274 (0.938–5.510) | – | |||
| Marital status | 0.123 | – | |||
| Single | 1.000 | – | |||
| Married | 1.283 (0.995–1.656) | – | |||
| Divorced/widowed/separated | 1.274 (0.768–2.113) | – | |||
| ICD-O-3/pathological subtype | 0.001* | 0.037* | |||
| 9180/3 osteosarcoma (NOS) | 1.000 | 1.000 | |||
| 9181/3 chondroblastic osteosarcoma | 0.968 (0.729–1.285) | 0.914 (0.680–1.228) | |||
| 9182/3 fibroblastic osteosarcoma | 0.811 (0.513–1.282) | 0.701 (0.439–1.121) | |||
| 9183/3 telangiectatic osteosarcoma | 0.800 (0.466–1.372) | 0.734 (0.424–1.273) | |||
| 9184/3 osteosarcoma in Paget disease of bone | 2.487 (0.796–7.771) | 1.659 (0.500–5.506) | |||
| 9185/3 small cell osteosarcoma | 0.290 (0.041–2.064) | 0.267 (0.037–1.924) | |||
| 9186/3 central osteosarcoma | 0.486 (0.240–0.984) | 0.540 (0.265–1.101) | |||
| 9187/3 intraosseous well-differentiated osteosarcoma | <0.001 (<0.001–8.807E+80) | <0.001 (<0.001–4.604E+114) | |||
| 9192/3 parosteal osteosarcoma | 0.149 (0.055–0.400) | 0.230 (0.082–0.645) | |||
| 9193/3 periosteal osteosarcoma | 0.118 (0.016–0.838) | 0.184 (0.026–1.320) | |||
| 9194/3 high-grade surface osteosarcoma | 1.897 (0.607–5.929) | 1.869 (0.593–5.887) | |||
| Primary site | 0.001* | 0.092 | |||
| Extremities | 1.000 | 1.000 | |||
| Pelvis/spine | 1.947 (1.328–2.856) | 1.248 (0.763–2.041) | |||
| Skull/mandible and others | 0.841 (0.577–1.225) | 0.667 (0.416–1.069) | |||
| Grade | <0.001* | 0.379 | |||
| Well/moderately differentiated; Grade I/II | 1.000 | 1.000 | |||
| Poorly/undifferentiated/anaplastic; Grade III/IV | 2.256 (1.593–3.194) | 1.279 (0.874–1.871) | |||
| Unknown | 1.689 (1.044–2.732) | 1.119 (0.669–1.871) | |||
| Laterality | 0.478 | – | |||
| Not a paired site (axial) | 1.000 | – | |||
| Left-origin of primary | 0.947 (0.649–1.383) | – | |||
| Right-origin of primary | 0.838 (0.571–1.231) | – | |||
| Bilateral, single primary | – | – | |||
| AJCC-T | <0.001* | 0.550 | |||
| T1 | 1.000 | 1.000 | |||
| T2 | 1.616 (1.281–2.038) | 1.146 (0.871–1.508) | |||
| T3 | 3.297 (1.946–5.584) | 0.966 (0.514–1.817) | |||
| AJCC–N | <0.001* | 0.002 | |||
| N0 | 1.000 | 1.000 | |||
| N1 | 6.698 (3.742–11.988) | 2.690 (1.443–5.017) | |||
| AJCC-M | <0.001* | <0.001* | |||
| M0 | 1.000 | 1.000 | |||
| M1 | 3.330 (2.614–4.242) | 2.848 (2.182–3.718) | |||
| Surgery code | <0.001* | 0.117 | |||
| Local or partial resection | 1.000 | 1.000 | |||
| Radical resection with limb salvage | 0.862 (0.624–1.191) | 0.763 (0.541–1.077) | |||
| Amputation | 1.322 (0.921–1.899) | 0.939 (0.634–1.391) | |||
| Forequarter/hindquarter/hemipelvectomy | 2.390 (1.459–3.915) | 1.310 (0.748–2.296) | |||
| Surgery, NOS | 1.432 (0.516–3.980) | 1.293 (0.456–3.670) | |||
| Radiation | <0.001* | 0.013* | |||
| None/unknown | 1.000 | 1.000 | |||
| Yes | 1.934 (1.363–2.745) | 1.646 (1.109–2.442) | |||
| Chemotherapy | 0.001* | 0.056 | |||
| No/unknown | 1.000 | 1.000 | |||
| Yes | 1.891 (1.307–2.736) | 1.518 (0.990–2.328) | |||
| Tumor size (mm) | 1.003 (1.002–1.004) | <0.001* | 1.002 (1.001–1.003) | <0.001* | |
| Extension (mm) | 1.024 (1.017–1.031) | <0.001* | 1.019 (1.011–1.028) | <0.001* | |
*, P<0.05. HR, hazard ratio; CI, confidence interval; AJCC, the American Joint Committee on Cancer; NOS, not otherwise specified.
The corresponding results defining the predictive factors for the analysis of OS were similar. Univariate analysis demonstrated that age, sex, marital status, pathological subtype (ICD-O-3 Hist/behave), primary site, grade, AJCC-T, AJCC-N, AJCC-M, tumor size, and tumor extension, surgery type, radiation, and chemotherapy were associated with OS (all P≤0.05), whereas race and laterality were not (P>0.05) (Table 3). Multivariate Cox analysis showed that older age, osteosarcoma NOS, AJCC-N1, AJCC-M1, radiation treatment, larger tumor size, and longer tumor extension were independent risk factors of OS (all P≤0.05) (Table 3).
Table 3
| Characteristics | Univariate analyses | Multivariate analyses | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (years) | 1.019 (1.013–1.024) | <0.001* | 1.034 (1.026–1.043) | <0.001* | |
| Sex | 0.002* | 0.195 | |||
| Male | 1.000 | 1.000 | |||
| Female | 0.718 (0.583–0.886) | 0.865 (0.695–1.077) | |||
| Race | 0.159 | – | |||
| White | 1.000 | – | |||
| Black | 0.901 (0.662–1.228) | – | |||
| Asian or Pacific Islander | 1.275 (0.905–1.797) | – | |||
| American Indian | 2.071 (0.855–5.016) | – | |||
| Marital status | 0.009 | 0.077 | |||
| Single | 1.000 | 1.000 | |||
| Married | 1.403 (1.105–1.781) | 0.851 (0.608–1.191) | |||
| Divorced/widowed/separated | 1.486 (0.943–2.341) | 0.515 (0.289–0.915) | |||
| ICD-O-3/pathological subtype | <0.001* | 0.029* | |||
| 9180/3 osteosarcoma (NOS) | 1.000 | 1.000 | |||
| 9181/3 chondroblastic osteosarcoma | 0.945 (0.718–1.242) | 0.879 (0.660–1.170) | |||
| 9182/3 fibroblastic osteosarcoma | 0.886 (0.582–1.348) | 0.821 (0.533–1.266) | |||
| 9183/3 telangiectatic osteosarcoma | 0.893 (0.546–1.460) | 0.780 (0.471–1.291) | |||
| 9184/3 osteosarcoma in Paget disease of bone | 3.003 (1.118–8.070) | 1.611 (0.565–4.594) | |||
| 9185/3 small cell osteosarcoma | 0.264 (0.037–1.883) | 0.251 (0.035–1.809) | |||
| 9186/3 central osteosarcoma | 0.505 (0.260–0.982) | 0.565 (0.289–1.107) | |||
| 9187/3 intraosseous well-differentiated osteosarcoma | <0.001 (<0.001–2.972E+77) | <0.001 (<0.001–1.000E+108) | |||
| 9192/3 parosteal osteosarcoma | 0.136 (0.051–0.365) | 0.210 (0.076–0.585) | |||
| 9193/3 periosteal osteosarcoma | 0.107 (0.015–0.763) | 0.150 (0.021–1.079) | |||
| 9194/3 high-grade surface osteosarcoma | 1.746 (0.559–5.453) | 1.762 (0.559–5.547) | |||
| Primary site | <0.001* | 0.075 | |||
| Extremities | 1.000 | 1.000 | |||
| Pelvis/spine | 2.145 (1.504–3.060) | 1.499 (0.949–2.367) | |||
| Skull/mandible and others | 1.026 (0.735–1.432) | 0.821 (0.535–1.258) | |||
| Grade | <0.001* | 0.141 | |||
| Well/moderately differentiated; Grade I/II | 1.000 | 1.000 | |||
| Poorly/undifferentiated/anaplastic; Grade III/IV | 2.256 (1.615–3.151) | 1.453 (1.004–2.102) | |||
| Unknown | 1.880 (1.202–2.942) | 1.406 (0.869–2.275) | |||
| Laterality | 0.165 | – | |||
| Not a paired site (axial) | 1.000 | – | |||
| Left-origin of primary | 0.790 (0.563–1.109) | – | |||
| Right-origin of primary | 0.718 (0.509–1.014) | – | |||
| Bilateral, single primary | – | – | |||
| AJCC-T | <0.001* | 0.785 | |||
| T1 | 1.000 | 1.000 | |||
| T2 | 1.447 (1.164–1.798) | 1.066 (0.821–1.382) | |||
| T3 | 2.799 (1.662–4.716) | 0.907 (0.487–1.691) | |||
| AJCC-N | <0.001* | 0.001* | |||
| N0 | 1.000 | 1.000 | |||
| N1 | 6.193 (3.463–11.073) | 2.835 (1.535–5.237) | |||
| AJCC-M | <0.001* | <0.001* | |||
| M0 | 1.000 | 1.000 | |||
| M1 | 3.096 (2.445–3.919) | 2.823 (2.178–3.659) | |||
| Surgery code | <0.001* | 0.107 | |||
| Local or partial resection | 1.000 | 1.000 | |||
| Radical resection with limb salvage | 0.755 (0.561–1.016) | 0.736 (0.535–1.014) | |||
| Amputation | 1.222 (0.876–1.705) | 0.961 (0.667–1.385) | |||
| Forequarter/hindquarter/hemipelvectomy | 2.050 (1.279–3.285) | 1.110 (0.647–1.906) | |||
| Surgery, NOS | 1.184 (0.429–3.265) | 0.936 (0.332–2.639) | |||
| Radiation | <0.001* | *0.037 | |||
| None/unknown | 1.000 | 1.000 | |||
| Yes | 1.974 (1.416–2.751) | 1.491 (1.024–2.170) | |||
| Chemotherapy | 0.006* | 0.174 | |||
| No/unknown | 1.000 | 1.000 | |||
| Yes | 1.587 (1.143–2.203) | 1.308 (0.888–1.928) | |||
| Tumor size (mm) | 1.003 (1.002–1.004) | <0.001* | 1.002 (1.001–1.003) | <0.001* | |
| Extension (mm) | 1.024 (1.018–1.030) | <0.001* | 1.020 (1.012–1.028) | <0.001* | |
*, P<0.05. HR, hazard ratio; CI, confidence interval; AJCC, the American Joint Committee on Cancer; NOS, not otherwise specified.
Establishment of the nomogram
Statistically significant factors from the results of the multivariate analyses were integrated to build the nomograms. To avoid the potential bias mentioned above, we excluded radiotherapy and chemotherapy in our final nomograms (18). Figure 2A,2B represents the nomograms for CSS and OS, respectively. As shown in Figure 2, the nomograms assign a score for each prognostic factor and by calculating the total points, the estimated CSS or OS at 1, 3 or 5 years can be obtained for individual patients. Higher total points correlate with worse patient survival.
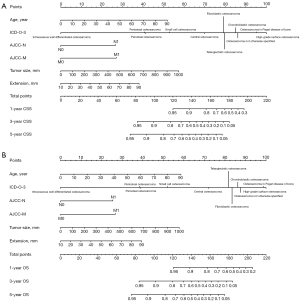
Evaluation and verification of the nomogram
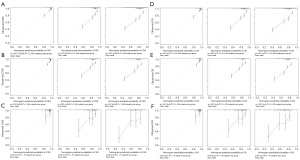
The verification of the nomograms was carried out in the training set and two validation sets. For CSS, the C-index of the training set was 0.731 (95% CI: 0.704–0.758); of the internal validation set was 0.713 (95% CI: 0.656–0.770); and of the external validation set was 0.721 (95% CI: 0.509–0.933). For OS, the C-index of the training set was 0.734 (95% CI: 0.708–0.759); of the internal validation set was 0.706 (95% CI: 0.651–0.761); and of the external validation set was 0.719 (95% CI: 0.503–0.935). These findings suggest that the discriminative ability was acceptable.
The calibration curves (Figure 3) for the training set (Figure 3A,3D), the internal verification set (Figure 3B,3E), and the external verification set (Figure 3C,3F), and for both CSS (Figure 3A-3C) and OS (Figure 3D-3F), were all close to the diagonal, indicating that the nomogram exhibited good concordance between the prediction and actual outcomes.
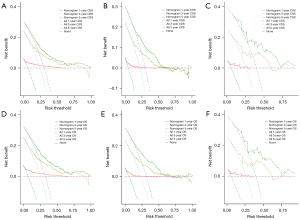
The DCA curves of the training and verification groups are shown in Figure 4. The graphics showed the ideal net benefit for CSS or OS at 1, 3 and 5 years, suggesting that the nomograms have good potential for clinical application.
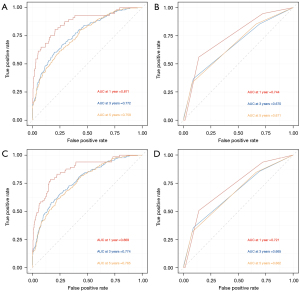
The tdROC analysis showed that the AUC values of our nomogram (in training set for the 12, 36 and 60 months) reached 0.871, 0.772, and 0.759 for CSS, while 0.869, 0.774, and 0.765 for OS. In the 6th AJCC TNM staging system, the AUC values of 0.744, 0.670, and 0.671 for CSS, and 0.721, 0.665, and 0.662 for OS (in training set for the 12, 36 and 60 months) were observed. The results revealed that the nomograms performed a better prediction for CSS and OS than the 6th AJCC TNM staging system (Figure 5A-5D).
Development of online webserver of our nomogram
An online nomogram can be accessed at “https://fengliwen.shinyapps.io/DNom-osteosarcoma-CSS/” and “https://fengliwen.shinyapps.io/DNom-osteosarcoma-OS/”. Predicted survival probabilities can be easily obtained by inputting clinicopathological characteristics and evaluating the generated nomograms.
Discussion
At present, the standard treatment mode for osteosarcoma is a surgical center multidisciplinary team approach, regardless of metastatic osteosarcoma (28,29). However, controversies remain about the stratification of risk in postoperative patients and affect the decision-making on the course of adjuvant therapy. In this study, a prediction nomogram for postoperative survival of osteosarcoma was established using a large sample of data from the SEER database; this nomogram can be used to estimate the prognosis of individual patients and guide the stratification of the disease risk. Using the multivariate Cox regression model, we found that older age, osteosarcoma NOS, AJCC-N1, AJCC-M1, radiation treatment, larger tumor size, and longer tumor extension were independently associated with worse survival outcome in patients with osteosarcoma. These findings remind clinicians that age, histological subtype, AJCC-Stage group, treatment strategies, tumor size, and tumor extension should be considered when surgery is performed.
Previous studies attributed a comparable poor prognosis in the presence of increasing age (8,30,31). Hagleitner et al. found that age is significantly associated with stage of presentation, tumor size, and histological response, and concluded that younger patients have a significantly better outcome than older patients (30). Tsuda et al. collected 110 patients with osteosarcoma over the age of 65 years, identified a significantly higher proportion of tumors arising in the trunk and metastases at diagnosis, and found that 5-year survival in this age group was only 32.7% (31). In other studies, evidence of age as an independent predictor of osteosarcoma survival remains controversial (32). A meta-analysis studied data from prospective neoadjuvant chemotherapy trials in osteosarcoma and the subgroup analysis did not indicate a statistically significant correlation between age and OS (P=0.12) (32). However, this meta-analysis divided age groups according to puberty and skeletal development, defined as follows: child (male, 0–12 years; female, 0–11 years); adolescent (male, 13–17 years; female, 12–16 years); adult (male, ≥18 years; female, ≥17 years), which may result in a loss of statistical power compared to the original continuous variable.
Regarding the histological subtype, our study suggested that osteosarcoma NOS was associated with poor survival compared to others. As shown in Tables 2,3, osteosarcoma in Paget disease of bone and high-grade surface osteosarcoma also appeared to suggest a poor prognosis (HR >1). Despite the low proportion, a high rate of metastasis was observed in previous reports (33,34). Deng et al. studied 23 patients with high-grade surface osteosarcoma; the 5-year OS rate was only 37.6% despite all patients were treated with a combination of surgery and systemic chemotherapy (34).
It is well established that patients with large tumors and longer tumor extension at initial presentation have an unfavorable prognosis (9,22). Various studies used different cutoff values for tumor size; Song et al. divided tumor size into three groups, which were <8, 8–13, and >13 cm (9); Dong et al. used the cutoff value of 6 and 12 cm (22). In our study, the original continuous data of tumor size and tumor extension were incorporated into the Cox analysis and a similar result was concluded, which further adds to the credibility that size and tumor extension are significant in predicting the prognosis of osteosarcoma.
Generally, osteosarcoma is considered relatively radioresistant, so radiotherapy is usually reserved for palliative treatment (35). Although radiotherapy did not improve survival in our study, it is evident that radiotherapy is valuable in terms of reducing pain intensity and local control when combined with chemotherapy (36). Currently, the beneficial role of neoadjuvant chemotherapy followed by surgical resection has been validated in osteosarcoma (4,5). Unexpectedly, univariate Cox regression showed that chemotherapy patients appear to be related to poor survival, although it did not reach statistical significance in multivariate analysis (Table 2). We evaluated the potential reasons associated with this result and found that chemotherapy use was associated with death risk, including Grade III/IV, fewer AJCC-Stage I, more AJCC-Stage IV, fewer AJCC-T1, more AJCC-M1, larger tumor size, and longer tumor extension, as shown in Table S2, thus this result may be influenced by other risk factors. Previous studies have also suggested that chemotherapy may lose effectiveness in specific populations (37). Iwata et al. investigated clinical characteristics and prognostic factors in patients older than 40 years with osteosarcoma, and found that chemotherapy contributed little to the prognosis in older osteosarcoma (37). Additionally, the radiotherapy and chemotherapy data were coded as ‘yes’ or ‘no/unknown’ in the SEER database; the result ‘no/unknown’, which indicated that there was no record of radiotherapy and chemotherapy in medical information, would introduce bias in survival analysis. Moreover, there were no detailed records of radiotherapy and chemotherapy regimens, which may have contributed to another bias. Therefore, we excluded treatment-related variables in our final nomograms.
It is worth mentioning that the most common location was the limbs, whose incidence proportion was over 80% in our study. Surgical treatment for limb osteosarcoma is usually divided into amputation and limb salvage. Although the techniques for limb salvage surgery are improving and its indications are expanding, it is widely accepted that limb salvage surgery should be recommended for patients if they are eligible for a safe surgical margin and respond well to chemotherapy (38). The effect of amputation and limb salvage on long-term survival is controversial. Bacci et al. studied 465 patients with limb salvage and 95 with amputation, and found no significant differences in 5-year OS between two groups; they concluded that limb salvage was relatively safe in osteosarcoma treated with neoadjuvant chemotherapy (38). A meta-analysis included ten studies and indicated that limb salvage was significantly superior to amputation according to 5-year OS; however, there appeared to be no significant difference according to local recurrence (39).
Several predictive models for osteosarcoma have been constructed in previous studies (9,26,40,41); however, the corresponding results will be more credible if they can be verified externally. In addition, whether they exhibit universal applicability in Asian/Chinese populations is also questionable. The present study used an Asian/Chinese population from a single center as a data source for external validation; the results of calibration curves, C-indexes, DCA, and AUC suggested that the nomogram had application value in China. To our knowledge, this is also the first nomogram with an online version for the prediction of osteosarcoma survival, which exhibits convenient applicability.
Limitations still exist in the present study. First, this study was a retrospective study, which is therefore susceptible to inherent bias. Second, due to some limitations of the SEER database, several biomarkers with potential clinical value in osteosarcoma, such as lactate dehydrogenase (42), alkaline phosphatase (43), and tumor necrosis rate/Huvos classification (7), were not included in this study. Third, even though recurrence and metastasis could also be endpoints for developing the corresponding nomograms (33,40), due to the insufficient number of such records in the SEER database, only survival predictions were made in the present study. Finally, we only enrolled patients who had undergone surgery; therefore, this nomogram is not applicable to patients who did not undergo surgery.
Conclusions
Using data from the SEER database, this study identified elder age, osteosarcoma NOS, AJCC-N1, AJCC-M1, radiation treatment, larger tumor size and longer extension as independent risk factors for CSS and OS of patients with osteosarcoma. Based on these independent prognostic variables, we constructed nomograms for predicting 1-, 3-, and 5-year OS and CSS. All the parameters of our nomograms showed acceptable performance, including calibration curves, C-indexes, DCA, and tdROC. This nomogram can provide individual risk assessment for the survival of patients with osteosarcoma who underwent surgery.
Acknowledgments
We thank the clinicians and institutions who shared their data to SEER database. We also thank Mr. Wei WANG (Jiangsu Province Hospital of Chinese Medicine) for his help and valuable discussions on statistics.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2756/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2756/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2756/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology [No. 2022 (0032)] and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res 2009;152:3-13. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Isakoff MS, Bielack SS, Meltzer P, et al. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J Clin Oncol 2015;33:3029-35. [Crossref] [PubMed]
- Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 2002;20:776-90. [Crossref] [PubMed]
- Senerchia AA, Macedo CR, Ferman S, et al. Results of a randomized, prospective clinical trial evaluating metronomic chemotherapy in nonmetastatic patients with high-grade, operable osteosarcomas of the extremities: A report from the Latin American Group of Osteosarcoma Treatment. Cancer 2017;123:1003-10. [Crossref] [PubMed]
- Kempf-Bielack B, Bielack SS, Jürgens H, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS). J Clin Oncol 2005;23:559-68. [Crossref] [PubMed]
- Bacci G, Longhi A, Versari M, et al. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer 2006;106:1154-61. [Crossref] [PubMed]
- Longhi A, Errani C, Gonzales-Arabio D, et al. Osteosarcoma in patients older than 65 years. J Clin Oncol 2008;26:5368-73. [Crossref] [PubMed]
- Song K, Song J, Chen F, et al. Prognostic nomograms for predicting overall and cancer-specific survival of high-grade osteosarcoma patients. J Bone Oncol 2018;13:106-13. [Crossref] [PubMed]
- Enneking WF, Spanier SS, Goodman MA. A system for the surgical staging of musculoskeletal sarcoma. 1980. Clin Orthop Relat Res 2003;4-18. [Crossref] [PubMed]
- Sheridan GA, Mellon L, Thompson EM, et al. Prognostic Importance of Pathological Fractures in Osteosarcomas. Ir Med J 2020;112:1021. [PubMed]
- Chen H, Liu J, Cheng Z, et al. Development and external validation of an MRI-based radiomics nomogram for pretreatment prediction for early relapse in osteosarcoma: A retrospective multicenter study. Eur J Radiol 2020;129:109066. [Crossref] [PubMed]
- Xu M, Dai N, Yang X, et al. Characteristics and prognosis of telangiectatic osteosarcoma: a population-based study using the Surveillance, Epidemiology and End Results (SEER) database. Ann Transl Med 2021;9:796. [Crossref] [PubMed]
- Zhang C, Guo X, Xu Y, et al. Lung metastases at the initial diagnosis of high-grade osteosarcoma: prevalence, risk factors and prognostic factors. A large population-based cohort study. Sao Paulo Med J 2019;137:423-9. [Crossref] [PubMed]
- Wang Z, Wu B, Zhou Y, et al. Predictors of the Survival of Primary and Secondary Older Osteosarcoma Patients. J Cancer 2019;10:4614-22. [Crossref] [PubMed]
- National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) program. Available online: http://seer.cancer.gov/data. Accessed December 22, 2018.
- Feng LW, Li J, Liang LF, et al. A Predictive Scoring System Based on Inflammatory and Tumor Markers for Gastric Cancer Patients Undergoing Curative Resection. Cancer Manag Res 2020;12:3937-48. [Crossref] [PubMed]
- Chen X, Pang Z, Wang Y, et al. The role of surgery for atypical bronchopulmonary carcinoid tumor: Development and validation of a model based on Surveillance, Epidemiology, and End Results (SEER) database. Lung Cancer 2020;139:94-102. [Crossref] [PubMed]
- American Joint Committee on Cancer: Cancer Staging Manual, 6th edition. New York: Springer, 2002.
- Pan Y, Chen D, Hu T, et al. Characteristics and Prognostic Factors of Patients With Osteosarcoma Older Than 60 Years From the SEER Database. Cancer Control 2019;26:1073274819888893. [Crossref] [PubMed]
- Qi L, Ren X, Liu Z, et al. Predictors and Survival of Patients with Osteosarcoma After Limb Salvage versus Amputation: A Population-Based Analysis with Propensity Score Matching. World J Surg 2020;44:2201-10. [Crossref] [PubMed]
- Dong Y, Wu W, Kang H, et al. Risk factors of regional lymph node (RLN) metastasis among patients with bone sarcoma and survival of patients with RLN-positive bone sarcoma. Ann Transl Med 2021;9:48. [Crossref] [PubMed]
- Evans DR, Lazarides AL, Visgauss JD, et al. Limb salvage versus amputation in patients with osteosarcoma of the extremities: an update in the modern era using the National Cancer Database. BMC Cancer 2020;20:995. [Crossref] [PubMed]
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. [Crossref] [PubMed]
- Li L, Greene T, Hu B. A simple method to estimate the time-dependent receiver operating characteristic curve and the area under the curve with right censored data. Stat Methods Med Res 2018;27:2264-78. [Crossref] [PubMed]
- Jiang Y, Wang T, Wei Z. Construction and Validation of Nomograms for Predicting the Prognosis of Juvenile Osteosarcoma: A Real-World Analysis in the SEER Database. Technol Cancer Res Treat 2020;19:1533033820947718. [Crossref] [PubMed]
- van Walraven C, Hawken S. Competing risk bias in Kaplan-Meier risk estimates can be corrected. J Clin Epidemiol 2016;70:101-5. [Crossref] [PubMed]
- García Franco CE, Torre W, Tamura A, et al. Long-term results after resection for bone sarcoma pulmonary metastases. Eur J Cardiothorac Surg 2010;37:1205-8. [Crossref] [PubMed]
- Farfalli GL, Albergo JI, Lobos PA, et al. Osteosarcoma lung metastases. Survival after chemotherapy and surgery. Medicina (B Aires) 2015;75:87-90. [PubMed]
- Hagleitner MM, Hoogerbrugge PM, van der Graaf WT, et al. Age as prognostic factor in patients with osteosarcoma. Bone 2011;49:1173-7. [Crossref] [PubMed]
- Tsuda Y, Ogura K, Shinoda Y, et al. The outcomes and prognostic factors in patients with osteosarcoma according to age: a Japanese nationwide study with focusing on the age differences. BMC Cancer 2018;18:614. [Crossref] [PubMed]
- Collins M, Wilhelm M, Conyers R, et al. Benefits and adverse events in younger versus older patients receiving neoadjuvant chemotherapy for osteosarcoma: findings from a meta-analysis. J Clin Oncol 2013;31:2303-12. [Crossref] [PubMed]
- Huang X, Zhao J, Bai J, et al. Risk and clinicopathological features of osteosarcoma metastasis to the lung: A population-based study. J Bone Oncol 2019;16:100230. [Crossref] [PubMed]
- Deng Z, Huang Z, Ding Y, et al. High-Grade Surface Osteosarcoma: Clinical Features and Oncologic Outcome. J Bone Oncol 2020;23:100288. [Crossref] [PubMed]
- DeLaney TF, Park L, Goldberg SI, et al. Radiotherapy for local control of osteosarcoma. Int J Radiat Oncol Biol Phys 2005;61:492-8. [Crossref] [PubMed]
- Lee JA, Paik EK, Seo J, et al. Radiotherapy and gemcitabine-docetaxel chemotherapy in children and adolescents with unresectable recurrent or refractory osteosarcoma. Jpn J Clin Oncol 2016;46:138-43. [PubMed]
- Iwata S, Ishii T, Kawai A, et al. Prognostic factors in elderly osteosarcoma patients: a multi-institutional retrospective study of 86 cases. Ann Surg Oncol 2014;21:263-8. [Crossref] [PubMed]
- Bacci G, Ferrari S, Lari S, et al. Osteosarcoma of the limb. Amputation or limb salvage in patients treated by neoadjuvant chemotherapy. J Bone Joint Surg Br 2002;84:88-92. [Crossref] [PubMed]
- He X, Gao Z, Xu H, et al. A meta-analysis of randomized control trials of surgical methods with osteosarcoma outcomes. J Orthop Surg Res 2017;12:5. [Crossref] [PubMed]
- Lu S, Wang Y, Liu G, et al. Construction and validation of nomogram to predict distant metastasis in osteosarcoma: a retrospective study. J Orthop Surg Res 2021;16:231. [Crossref] [PubMed]
- Jiang J, Pan H, Li M, et al. Predictive model for the 5-year survival status of osteosarcoma patients based on the SEER database and XGBoost algorithm. Sci Rep 2021;11:5542. [Crossref] [PubMed]
- Chen J, Sun MX, Hua YQ, et al. Prognostic significance of serum lactate dehydrogenase level in osteosarcoma: a meta-analysis. J Cancer Res Clin Oncol 2014;140:1205-10. [Crossref] [PubMed]
- Kim SH, Shin KH, Moon SH, et al. Reassessment of alkaline phosphatase as serum tumor marker with high specificity in osteosarcoma. Cancer Med 2017;6:1311-22. [Crossref] [PubMed]


