
miR-135a inhibits the proliferation of HBV-infected hepatocellular carcinoma cells by targeting HOXA10
Highlight box
Key findings
• HBV promoted the proliferation and invasion and inhibited the apoptosis of HCC cells via the miR-135a/HOXA10 pathway.
What is known and what is new?
• miRNAs regulate HCC cell proliferation, apoptosis, migration and invasion.
• This study explored the mechanism and roles of miR-135a and HOXA10 in HBV-induced HCC.
What are the implications, and what should change now?
• This study provides a theoretical basis and new targets for further research on the mechanism and clinical treatment of HBV-induced HCC.
Introduction
At present, the incidence of hepatocellular carcinoma (HCC) is very high (1,2). Hepatitis B virus (HBV) infection is the main cause of type B HCC onset. Globally, 55% of HCC patients are infected with HBV, while in Asia and Africa, this proportion is as high as 80% (3). Adults with chronic hepatitis B will eventually develop cirrhosis and/or HCC. Cirrhosis and HCC were responsible for 887,000 HBV deaths in 2015 (4). HBV is an easily mutated virus with genetic diversity. There are more than 9 genotypes of HBV. Different genotypes of HBV have different prevalence, geographic distribution, natural history, disease progression, and treatment outcomes (5). Although the specific mechanism through which the HBV virus induces hepatocellular malignancies is still not very clear, some studies have reported that the various intracellular regulatory mechanisms involved in HOXA10 are closely related to the occurrence and development of various types of cancers (6,7). HOXA10 is a member of the homeobox gene family and is overexpressed in cancer tissues. Studies have found that HOXA10 is highly expressed in oral cancer (8), endometrial cancer (9), breast cancer (10), and other tissues, which is related to the grade and invasion of the corresponding tumors. On the one hand, in normal liver cells infected with the HBV virus, the HOXA10 protein promotes the replication of the HBV viral genome (7). On the other hand, it exerts a carcinogenic effect by disturbing the epigenetic modification of the host cells (11). The HOXA10 protein can promote the expression of oncogenes and inhibit the activation of tumor suppressor genes by changing the ribosomal histone modification of cells or via DNA methylation modification (12). Importantly, HOXA10 can inhibit the proliferation of HCC cells through HDAC1 and induce cell cycle arrest and apoptosis (6).
Recent study has reported that HOXA10 is related to the expression of certain microRNA (miRNA) in liver cells (11). MiRNA is a type of endogenous non-coding small RNA with complex regulatory functions found in eukaryotic cells (13). It regulates the expression of target genes at the post-transcriptional level and participates in the regulation of many cell biological processes (14). Studies have shown that a variety of miRNAs play important roles in the occurrence and development of HCC caused by HBV infection (15). Exosome miR-142-3p secreted by HBV-HCC cells can regulate ferroptosis and HCC progression in M1-type macrophages by targeting SLC3A2 (16). MiR-744-5p has been shown to regulate the behavior of HCC cells by regulating the TGF-β1 signaling pathway and epithelial-mesenchymal-transition (EMT) (17). Although the effects of HOXA10 on the expression of miRNA in cells have been reported, the regulatory roles between miR-135a and HOXA10 in terms of biological function remain unknown. It has been reported that miR-135a is up-regulated in HCC (18). Our previous study confirms inhibiting miR-135a expression could promote HCC cell apoptosis (19). Therefore, miR-135a might be significantly related to the occurrence and development of HCC. At present, studies have not reported on the effects of HBV on the changes in the overall expression profile of miRNAs in liver cells or the bioinformatics analysis of the cellular pathways involved in these miRNAs.
This study explored the mechanism and roles of miR-135a and HOXA10 in HBV-indued HCC and provided a theoretical basis and new targets for further research on the mechanism and clinical treatment of HCC induced by HBV viral infection. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2789/rc).
Methods
Sample collection
The HCC tissues used in this study were obtained from 90 patients with HCC, including 80 males and 10 females, with an average age of 52.37 years. Nineteen cases were HBV negative, and 71 cases were HBV positive. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of Xiangshan First People’s Hospital, Ningbo Fourth Hospital (No. 20190530-2), and all subjects voluntarily participated in this study and signed the written informed consent.
Cell culture and transfection
Human normal hepatocytes (LO2), hepatoma cells (Bel-7402, Huh7, HepG2), and HBV-infected hepatoma cells (HepG2.2.15) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium containing 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA), 100 µL/mL penicillin, and 0.1 mg/mL streptomycin in a 5% CO2 incubator at 37 °C.
Transfection was performed when the cell confluence reached 70–80%, and Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used for transient transfection according to the manufacturer’s instructions. After 6 h, the old medium was discarded and replaced by the fresh complete medium. After culturing for 24 h, the cells were used for in vitro experiments.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The total RNAs of cells in each group were extracted using Trizol reagent, and complementary DNA (cDNA) was synthesized by reverse transcription according to the previous literature (20,21). The expression level of miR-135a was determined using the TaqMan miRNA reverse transcription kit (Life Technologies, USA). Using U6 as the internal parameter, the expression level of miR-135a in each group was calculated according to the 2 −△△CT method.
Dual luciferase experiment
The wild-type and mutant luciferase plasmids of HOXA10 were constructed and co-transfected with a miR-135a mimics and inhibitor. Luciferase activity was detected using a dual luciferase assay kit (Promega, WI, USA). The luminous intensity of each well was measured with a microplate fluorometer (Thermo Scientific, USA), and the dual luciferase activity of each group was calculated. Transfection was performed in duplicate and repeated three times.
Cell Counting Kit-8 (CCK)-8 assay
1×103 to 3×103 cells were inoculated in 96-well plates and cultured overnight. The cells were treated according to the experimental requirements. After the cell culture reached the target time, the medium was discarded and replaced by the fresh medium containing 10% CCK-8 reagent (Meilun, Dalian, China). After incubation for 1–4 h, the absorbance value of optical density (OD450) was measured using a multifunctional microplate reader (Thermo Scientific, USA).
Apoptosis assay
Cells from different groups were collected in a flow tube. Next, the cells were digested with 1 mL of 0.25% trypsin. The cell suspension was collected in a flow tube for 300 g centrifugation for 5 min and the supernatant was discarded. The pelleted cells were then resuspended in 300 µL of binding buffer. Subsequently, 5 µL Annexin V-fluorescein isothiocyanate (FITC) was added, mixed, and incubated in the dark for 10 min. Next, 5 µL of prodium iodide (PI) solution was added and the cells were incubated in the dark for 5 min after mixing. Finally, the apoptosis rate was detected by flow cytometry within 1 h.
Transwell assay
First, 600 µL of medium containing 10% FBS was added to the lower chamber of the transwell plate, and 5×104 cells were inoculated in the upper chamber and maintained in the incubator for 24 h. The liquid in the chamber was then sucked up and fixed in the hole pre-filled with about 800 µL of methanol at room temperature for 30 min. Subsequently, the cells were stained with 800 µL of Giemsa staining solution for 30 min at room temperature, and then gently rinsed several times with phosphate buffer saline (PBS) and observed under the microscope (Nikon TiU, Japan).
Western blotting assay
The total cell protein was extracted using protein lysate and the protein concentration was calculated as previously described (22-24). For Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) experiments, the same amount of protein solution was added to each well. After electrophoresis, the protein was transferred to a polyvinylidene fluoride (PVDF) membrane and blocked with 5% skim milk at room temperature for 2 h. The primary antibody was incubated overnight at 4 °C. After the membrane was washed three times with Tris Buffered Saline Tween (TBST), the secondary antibody was incubated for 1 h at room temperature. After the membrane was washed three times with TBST, enhanced chemiluminescence (ECL) luminescent solution was used to analyze the protein gray value. All antibodies were purchased from Cell Signaling Technology, USA. The dilution ratio of the primary antibody was 1:1,000, and the dilution ratio of the secondary antibody was 1:4,000.
Statistical analysis
The data were analyzed using GraphPad Prism 7.0 software (GraphPad Software, USA). The data were expressed as means ± standard deviation (SD). The comparisons among multiple groups were performed using one-way analysis of variance (ANOVA) followed by the post hoc test for multiple comparisons. P<0.05 indicated a significant difference.
Results
miR-135a expression in HCC tissues and cells
HBV can seriously aggravate the prevalence of liver cancer patients, and it is very important to effectively study HCC disease under HBV infection. To explore the expression of miR-135a in HCC, we detected the expression level of miR-135a in HCC tissues and cells using qRT-PCR analysis. The results indicated that the expression level of miR-135a was significantly decreased in the tissues from the HBV-positive group, as compared with those from the HBV-negative group (Figure 1A). Also, the results from subsequent experiments showed that the expression of miR-135a was significantly reduced in Bel-7402, Huh7, HepG2, and HepG2.2.15 cells compared to LO2 cells, while the expression level was the lowest in the HepG2.2.15 cells (Figure 1B). These results indicated that miR-135a was decreased in HCC tissues and cells, which suggested that miR-135a might be related to the occurrence and development of HCC.

The effects of miR-135a on the cell proliferation of HCC cells
The expression of miR-135a was significantly decreased in HepG2 and HepG2.2.15 cells. Thus, these HCC cells (HepG2 and HepG2.2.15) were selected for further experiments. These experiments were used to explore the effects of miR-135a on cell proliferation through CCK-8 assay and by detecting the expression of proliferation-related proteins. The CCK-8 assay revealed that the OD values of the miR-135a mimics + HepG2 group and the HepG2.2.15 + miR-135a mimics group were markedly reduced compared with the HepG2 and HepG2.2.15 group (Figure 2A). Moreover, the OD values of the HepG2.2.15 + miR-135a mimics group were the lowest (Figure 2A). These findings suggested that miR-135a significantly inhibited the proliferation of HCC cells, and the proliferation rate in HBV-infected cells was also inhibited (Figure 2A).

Also, the expression levels of proliferation-related proteins, proliferating cell nuclear antigen (PCNA) and CyclinD1, were significantly downregulated in the miR-135a mimics + HepG2 group and the HepG2.2.15 + miR-135a mimics group, as compared with the HepG2 and HepG2.2.15 group (Figure 2B). Furthermore, the expression levels of PCNA and CyclinD1 in the HepG2.2.15 + miR-135a mimics group were the lowest (Figure 2B). These results indicated that miR-135a inhibited HCC cell proliferation, especially after HBV infection.
The effects of miR-135a on the cell invasion of HCC cells
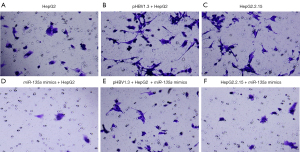
To further investigate the roles of miR-135a in the development of HCC, we determined the HCC cell invasion after miR-135a transfection. Transwell assays were applied to detect the invasion ability of miR-135a on HCC cells (Figure 3). According to the experimental results, the number of invasive cells in the miR-135a mimics group was significantly decreased compared to the untransfected miR-135a group (Figure 3A-3F). Interestingly, compared with the pHBV1.3 + HepG2 group, the number of invasive cells in the pHBV1.3 + HepG2 + miR-135a mimics group was significantly reduced (Figure 3A-3F). Additionally, the number of invasive cells in the HepG2.2.15 + miR-135a mimics group was lower than that in the HepG2.2.15 group (Figure 3A-3F). Taken together, these results showed that the overexpression of miR-135a significantly inhibited the invasion ability of HCC cells, especially after HBV infection.
The effects of miR-135a on the apoptosis of HCC cells
We further detected the roles of miR-135a in the apoptosis of HCC cells to determine the functions of miR-135a in HCC. Through flow cytometry experiments, we found that the miR-135a mimics + HepG2 group and the HepG2.2.15 + miR-135a mimics group both increased the apoptosis rate of HCC cells, as compared to the HepG2 group (Figure 4A). Also, the apoptosis rate of the HepG2.2.15 + miR-135a mimics group was significantly higher than that of the HepG2.2.15 group (Figure 4A). These results suggested that miR-135a markedly increased the apoptotic rate of HCC cells, and the level of apoptosis was much higher under HBV infection (Figure 4A).

In addition, the results of western blotting experiments also showed that the expressions of pro-apoptotic proteins (Bax and Cleaved Casepase-3) were upregulated in HCC cells following miR-135a mimics transfection (Figure 4B). Meanwhile, the expression of the anti-apoptotic protein Bcl-2 was markedly downregulated in HCC cells transfected with miR-135a mimics (Figure 4B). These results suggested that the overexpression of miR-135a promoted the apoptosis of HCC cells under HBV infection.
HOXA10 was the target of miR-135a
To further investigate the mechanism of miR-135a in HCC cells, the target genes of miR-135a were further determined. Through the dual luciferase experiment, we found that the miR-135a mimics and the WT-HOXA10-3’UTR plasmid group had significantly decreased luciferase activity compared with the WT-HOXA10-3’UTR plasmid and mimics negative control (NC) co-transfection group (Figure 5A). However, compared with the MUT-HOXA10-3’UTR plasmid and mimics NC co-transfection group, the luciferase activity of the miR-135a mimics and the MUT-HOXA10-3’UTR plasmid group did not exhibit a notable change (Figure 5A).
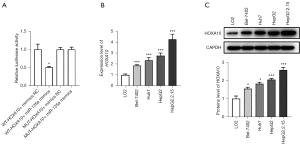
HOXA10, as a key cancer gene, plays an important role in liver cancer. Therefore, we examined the expression level of HOXA10 in HCC cells. The results showed that the expression level of HOXA10 was significantly increased in HCC cells, especially in the HBV-transfected HepG2 cells, as compared with LO2 cells (Figure 5B). Also, the western blotting results showed that the expression level of HOXA10 in HCC cells was significantly increased (Figure 5C). The above results suggested that HOXA10 was significantly upregulated in HCC cells, especially after HBV infection, and thus, HOXA10 might be a target of miR-135a in HCC cells.
The effects of HOXA10 on cell proliferation in HCC cells
A CCK-8 assay was applied to detect the roles of HOXA10 in HCC cells. The CCK-8 experiment showed that miR-135a mimics and si-HOXA10 could inhibit the proliferation of HCC cells, and co-transfection of the miR-135a mimics and si-HOXA10 could significantly reduce the OD values of HCC cells in the HepG2 + pHBV1.3 cell group (Figure 6A). Similarly, in the HepG2.2.15 cell group, co-transfection of the miR-135a mimics and si-HOXA10 also substantially inhibited the proliferation of HCC cells (Figure 6B).

The protein expression levels of proliferation-related proteins, PCNA and CyclinD1, were further measured using a western blot assay. The results indicated that in HepG2 cells transfected with pHBV1.3 or HepG2.2.15 cells, the miR-135a mimics + si-HOXA10 considerably reduced the expression levels of PCNA and CyclinD1 (Figure 6C,6D). Together, these results showed that miR-135a might inhibit the proliferation of HCC cells through its target gene, HOXA10.
The effects of HOXA10 on cell invasion of HCC cells
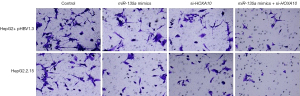
Our results showed that miR-135a could inhibit the invasion of HCC cells. We then investigated the roles of HOXA10 in HCC cell invasion. By silencing the target gene, HOXA10, we found that the number of invasive cells in miR-135a mimics + si-HOXA10 was significantly reduced in the HepG2 + pHBV1.3 cell group (Figure 7). Similarly, in the HepG2.2.15 cell group, the number of invasive cells in miR-135a mimics + si-HOXA10 was also markedly reduced (Figure 7). Therefore, these results suggested that miR-135a could significantly inhibit the invasion of HCC cells by silencing HOXA10 in HBV-infected HCC cells.
The effect of HOXA10 on the cell apoptosis of HCC cells
In the HepG2 + pHBV1.3 group, the co-transfection of miR-135a mimics and si-HOXA10 significantly increased the apoptotic rate of HCC cells, and the same results were also obtained in the HepG2.2.15 group (Figure 8A,8B). Furthermore, the western blot assay results showed that the miR-135a mimics and si-HOXA10 co-transfection also upregulated the protein expression level of the pro-apoptotic proteins, Bax and Cleaved Caspase-3, and downregulated the protein expression of the anti-apoptotic protein, Bcl-2, in the HepG2 + pHBV1.3 group (Figure 8C). In addition, the same results were also obtained in the HepG2.2.15 group (Figure 8D). The above results indicated that miR-135a could significantly promote the apoptosis of HCC cells by silencing HOXA10 in HBV-infected HCC cells.

The effects of miR-135a/HOXA10 on the activation of the MAPK pathway
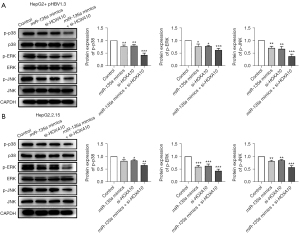
The MAPK pathway is a classic inflammation pathway, and the inflammatory response is a protective mechanism for the body to resist the invasion of pathogenic microorganisms. Thus, the MAPK pathway might be related to the miR-135a and HOXA10-mediated regulatory roles in HCC cells and further experiments were performed to investigate this hypophysis. The western blot analysis results showed that, compared with the control group, the miR-135a mimics + si-HOXA10 group significantly reduced the expression levels of phosphorylated proteins the p-p38, p-ERK, and p-JNK in HepG2 cells transfected with pHBV1.3 (Figure 9A). In HepG2.2.15 cells, the expression levels of p-p38, p-ERK, and p-JNK were also significantly inhibited via the co-transfection of miR-135a and si-HOXA10 (Figure 9B). Therefore, miR-135a/si-HOXA10 could significantly inhibit the activation of the MAPK pathway, which suggested that miR-135a might regulate the development of HCC by inhibiting the activation of the MAPK pathway.
Discussion
HCC has a high incidence and poor prognosis. Various types of genes and signal pathways are dysregulated in the pathogenesis of HCC (25). Recent studies have shown that more than 50% of miRNAs are located in relevant oncogenomic regions, and their expression is frequently dysregulated in cancer, highlighting their key role in tumorigenesis (13,26). Currently, miRNA mimics and siRNA, as targeted molecular therapy programs, are favored by the medical community (27). MiRNAs have proved to be dysregulated in HBV-infected HCC. 37 miRNAs are differently expressed in HBV-related TCGA-HCC cohorts between TPX2 Microtubule Nucleation Factor (TPX2)low and TPX2high groups (28). MiR-933 could affect cell growth and HBV DNA synthesis (29). LINC01232 could sponge miR-708-5p to regulate HCC progression (30). MiR-135a has been confirmed to play an important role in the invasion and metastasis of a variety of tumors. For example, it promotes the growth and invasion of colorectal cancer through metastasis inhibitor 1 in vitro (31), and inhibits the proliferation of gastric cancer cells by targeting JAK2 (32). However, there are few studies on miR-135a in HBV-infected liver tumors. Therefore, it is of practical significance to study the effect of miR-135a on HCC under HBV infection.
To explore the relationship between HBV and miR-135a, we used qRT-PCR to verify whether HBV could regulate the expression of miR-135a. The results showed that the expression of miR-135a was significantly down-regulated in HCC tissues and cells. This suggests that HBV may downregulate the expression of miR-135a through a particular mechanism. So, we attempted to determine how miR-135a is regulated by HBV. It is known that the biosynthesis of miRNAs is regulated by numerous aspects and levels, including the transcription level of miRNAs, RNA modification processes such as RNA methylation, and the localization of Ago proteins, etc. Among these, abnormal regulation of the miRNA transcription level is an important factor for the abnormal expression of miRNA. The abnormal regulation of miR-135a determines the fate of hepatoma cells to a certain extent. In our research, we observed that miR-135a is lowly expressed in HCC tissues and cell lines, and this downregulation suppressed the proliferation of HCC cells.
Invasion ability and apoptosis level are important indicators reflecting the vitality of HCC cells (33). It is known that HCC cells exert a strong ability to quickly invade surrounding cells, resulting in cell pathology and death (34,35). MiRNA regulation of tumor cells is inseparable from its influence on the invasion ability of tumor cells. A study has shown that miR-135a promotes the migration and invasion of HCC cells by targeting fork head box O1 (36). In this study, we came to the opposite conclusion; miR-135a not only does not promote the invasion ability of HepG2 or HepG2.2.15 cells but also inhibits their invasion, which is the more interesting aspect. MiRNA is known to possess the ability of two-way regulation, both promoting and inhibiting, so this may happen. By detecting the apoptosis ability, we found that under HBV infection, miR-135a significantly promoted the apoptosis of HCC cells, and at the same time elevated the expression of related pro-apoptotic proteins and reduced the expression of related anti-apoptotic proteins. These results illustrated the inhibitory effect exerted by HBV on the apoptosis of HCC cells, one of which was achieved by regulating the expression of miR-135a.
HOXA10 is a human homeobox gene; it is a DNA-binding transcription factor with sequence-specific DNA-binding activity (37). It is widely involved in reproductive tract development, embryo implantation regulation, and cell-directed differentiation and proliferation (38). HOXA10 plays an important role in embryonic development, differentiation, cancer, and hematopoietic diseases. Studies have shown that HOXA10 promotes gastric cancer metastasis by partially mediating epithelial-mesenchymal transition via regulation of the TGFB2/Smad/METTL3 signaling axis (39). LncHOXA10 drives the self-renewal and tumorigenesis of hepatic TICs through transcriptional activation of HOXA10 (40). However, so far, the role of HOXA10 in HBV infection has been rarely reported. Zhu et al. (41) used gene chip screening and found that HOXA10 was highly expressed in HepG2.2.15 cell lines, which integrated the entire HBV genome.
The results of the experiments in this study confirmed that HOXA10 was the target gene of miR-135a. Moreover, the high expression of miR-135a and silencing of HOXA10 exerted a similar effect on HBV-infected HCC cells. It is known that miRNA plays a role by regulating target mRNA, so miR-135a might exert a regulatory function on HCC cells by regulating the expression of HOXA10. Importantly, the high expression of miR-135a and silencing of HOXA10 could significantly enhance the unilateral effects of both on HBV infection of HCC cells.
In addition, this study investigated the MAPK pathway regulation by miR-135a/HOXA10. MAPK is a classic inflammatory pathway that plays an important role in the occurrence and development of tumors. Studies have shown that isoquercitrin inhibits the progression of liver cancer in vivo and in vitro by regulating the MAPK signaling pathway (42). Interestingly, miRNAs and MAPK exhibit significant associations in the occurrence and development of tumor diseases, and jointly affect disease progression. MiR-101 can affect the proliferation of hepatoma cells through the MAPK/ERK signaling pathway (43). In addition, miR-338-3p overexpression reduces the expression of MACC1 in cervical cancer cells via the MAPK pathway, and significantly inhibits the proliferation of cervical cancer cells and induces apoptosis (44). Although miRNAs can participate in the development of liver cancer by regulating the MAPK pathway, it is still unknown whether miR-135a regulates the progression of liver cancer through the MAPK pathway. In this study, we found that miR-135a could significantly inhibit MAPK pathway activation.
Conclusions
In conclusion, HBV promoted the proliferation and invasion of HCC cells and inhibited the apoptosis by regulating miR-135a/HOXA10 pathway. Importantly, miR-135a also promotes cell proliferation and inhibits MAPK pathway activation. This study provides a theoretical basis for understanding the mechanism of HBV infection of HCC cells.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2789/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2789/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2789/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of Xiangshan First People’s Hospital, Ningbo Fourth Hospital (No. 20190530-2), and all subjects voluntarily participated in this study and signed the written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang DX, Yang X, Lin JZ, et al. Efficacy and safety of lenvatinib for patients with advanced hepatocellular carcinoma: A retrospective, real-world study conducted in China. World J Gastroenterol 2020;26:4465-78. [Crossref] [PubMed]
- Li Y, Meng L, Zhao B. The roles of N6-methyladenosine methylation in the regulation of bone development, bone remodeling and osteoporosis. Pharmacol Ther 2022;238:108174. [Crossref] [PubMed]
- Xie Y, Hepatitis B. Virus-Associated Hepatocellular Carcinoma. Adv Exp Med Biol 2017;1018:11-21. [Crossref] [PubMed]
- Global Burden of Disease Liver Cancer Collaboration. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol 2017;3:1683-91. [Crossref] [PubMed]
- Liu Y, Park D, Cafiero TR, et al. Molecular clones of genetically distinct hepatitis B virus genotypes reveal distinct host and drug treatment responses. JHEP Rep 2022;4:100535. [Crossref] [PubMed]
- Zhang Y, Chen J, Wu SS, et al. HOXA10 knockdown inhibits proliferation, induces cell cycle arrest and apoptosis in hepatocellular carcinoma cells through HDAC1. Cancer Manag Res 2019;11:7065-76. [Crossref] [PubMed]
- Yang Q, Zhang Q, Zhang X, et al. HoxA10 Facilitates SHP-1-Catalyzed Dephosphorylation of p38 MAPK/STAT3 To Repress Hepatitis B Virus Replication by a Feedback Regulatory Mechanism. J Virol 2019;93:e01607-18. [Crossref] [PubMed]
- Yan X, Cong B, Chen Q, et al. Silencing lncRNA HOXA10-AS decreases cell proliferation of oral cancer and HOXA10-antisense RNA can serve as a novel prognostic predictor. J Int Med Res 2020;48:300060520934254. [Crossref] [PubMed]
- Zhang L, Wan Y, Jiang Y, et al. Upregulation HOXA10 homeobox gene in endometrial cancer: role in cell cycle regulation. Med Oncol 2014;31:52. [Crossref] [PubMed]
- Mustafa M, Lee JY, Kim MH. CTCF negatively regulates HOXA10 expression in breast cancer cells. Biochem Biophys Res Commun 2015;467:828-34. [Crossref] [PubMed]
- Xiao ZD, Jiao CY, Huang HT, et al. miR-218 modulate hepatocellular carcinoma cell proliferation through PTEN/AKT/PI3K pathway and HoxA10. Int J Clin Exp Pathol 2014;7:4039-44. [PubMed]
- Shao L, Chen Z, Peng D, et al. Methylation of the HOXA10 Promoter Directs miR-196b-5p-Dependent Cell Proliferation and Invasion of Gastric Cancer Cells. Mol Cancer Res 2018;16:696-706. [Crossref] [PubMed]
- Gao M, Zhang Z, Sun J, et al. The roles of circRNA-miRNA-mRNA networks in the development and treatment of osteoporosis. Front Endocrinol (Lausanne) 2022;13:945310. [Crossref] [PubMed]
- Zou J, Sun J, Chen H, et al. The regulatory roles of miR-26a in the development of fracture and osteoblasts. Ann Transl Med 2022;10:37. [Crossref] [PubMed]
- Lou W, Liu J, Ding B, et al. Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC. J Transl Med 2019;17:7. [Crossref] [PubMed]
- Hu Z, Yin Y, Jiang J, et al. Exosomal miR-142-3p secreted by hepatitis B virus (HBV)-hepatocellular carcinoma (HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression. J Gastrointest Oncol 2022;13:754-67. [Crossref] [PubMed]
- Huang W, Chen Q, Dai J, et al. miR-744-5p suppresses tumor proliferation and metastasis by targeting transforming growth factor-beta 1 (TGF-β1) in hepatocellular carcinoma (HCC). J Gastrointest Oncol 2021;12:1811-22. [Crossref] [PubMed]
- Deng X, Cheng J, Zhan N, et al. MicroRNA-135a expression is upregulated in hepatocellular carcinoma and targets long non-coding RNA TONSL-AS1 to suppress cell proliferation. Oncol Lett 2021;22:808. [Crossref] [PubMed]
- Lin J, Lian X, Xue S, et al. HBV Promotes the Proliferation of Liver Cancer Cells through the hsa_circ_0000847/miR-135a Pathway. Evid Based Complement Alternat Med 2022;2022:7332337. [Crossref] [PubMed]
- Li Y, Zou J, Li B, et al. Anticancer effects of melatonin via regulating lncRNA JPX-Wnt/beta-catenin signalling pathway in human osteosarcoma cells. J Cell Mol Med 2021;25:9543-56. [Crossref] [PubMed]
- Yan G, Zhang L, Feng C, et al. Blue light emitting diodes irradiation causes cell death in colorectal cancer by inducing ROS production and DNA damage. Int J Biochem Cell Biol 2018;103:81-8. [Crossref] [PubMed]
- Ye D, Bao Z, Yu Y, et al. Inhibition of cardiomyocyte differentiation of human induced pluripotent stem cells by Ribavirin: Implication for its cardiac developmental toxicity. Toxicology 2020;435:152422. [Crossref] [PubMed]
- Han Z, Yu Y, Xu J, et al. Iron Homeostasis Determines Fate of Human Pluripotent Stem Cells Via Glycerophospholipids-Epigenetic Circuit. Stem Cells 2019;37:489-503. [Crossref] [PubMed]
- Zhang Z, Ha SH, Moon YJ, et al. Inhibition of SIRT6 potentiates the anti-tumor effect of doxorubicin through suppression of the DNA damage repair pathway in osteosarcoma. J Exp Clin Cancer Res 2020;39:247. [Crossref] [PubMed]
- Tsuchida T, Lee YA, Fujiwara N, et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol 2018;69:385-95. [Crossref] [PubMed]
- Wei K, Ma Z, Yang F, et al. M2 macrophage-derived exosomes promote lung adenocarcinoma progression by delivering miR-942. Cancer Lett 2022;526:205-16. [Crossref] [PubMed]
- Zhang SG, Liu CY, Li L, et al. Examination of artificial MiRNA mimics with centered-site complementarity for gene targeting. PLoS One 2013;8:e72062. [Crossref] [PubMed]
- Li G, Wang Z, Chen D, et al. Comprehensive analysis of a TPX2-related TRHDE-AS1/PKIA ceRNA network involving prognostic signatures in Hepatitis B virus-infected hepatocellular carcinoma. Front Cell Infect Microbiol 2022;12:1025900. [Crossref] [PubMed]
- Cheng Y, Shi W, Cui X, et al. Long Noncoding RNA TFAP2A-AS1 Suppressed Hepatitis B Virus Replication by Modulating miR-933/HDAC11. Dis Markers 2022;2022:7733390. [Crossref] [PubMed]
- Guo L, Gao S, Sun W, et al. Elevated LINC01232 is associated with poor prognosis and HBV infection in hepatocellular carcinoma patients and contributes to tumor progression in vitro. Clin Res Hepatol Gastroenterol 2022;46:101813. [Crossref] [PubMed]
- Zhou W, Li X, Liu F, et al. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin (Shanghai) 2012;44:838-46. [Crossref] [PubMed]
- Wu H, Huang M, Cao P, et al. MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol Ther 2012;13:281-8. [Crossref] [PubMed]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. [Crossref] [PubMed]
- Singh V, Yeoh BS, Chassaing B, et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018;175:679-694.e22. [Crossref] [PubMed]
- Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev 2017;121:27-42. [Crossref] [PubMed]
- Zeng YB, Liang XH, Zhang GX, et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int 2016;16:63. [Crossref] [PubMed]
- Eklund EA, Goldenberg I, Lu Y, et al. SHP1 protein-tyrosine phosphatase regulates HoxA10 DNA binding and transcriptional repression activity in undifferentiated myeloid cells. J Biol Chem 2002;277:36878-88. [Crossref] [PubMed]
- Guo C, Ju QQ, Zhang CX, et al. Overexpression of HOXA10 is associated with unfavorable prognosis of acute myeloid leukemia. BMC Cancer 2020;20:586. [Crossref] [PubMed]
- Song C, Zhou C. HOXA10 mediates epithelial-mesenchymal transition to promote gastric cancer metastasis partly via modulation of TGFB2/Smad/METTL3 signaling axis. J Exp Clin Cancer Res 2021;40:62. [Crossref] [PubMed]
- Shao M, Yang Q, Zhu W, et al. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol Cancer 2018;17:173. [Crossref] [PubMed]
- Zhu C, Song H, Xu F, et al. Hepatitis B virus inhibits the expression of complement C3 and C4, in vitro and in vivo. Oncol Lett 2018;15:7459-63. [Crossref] [PubMed]
- Huang G, Tang B, Tang K, et al. Isoquercitrin inhibits the progression of liver cancer in vivo and in vitro via the MAPK signalling pathway. Oncol Rep 2014;31:2377-84. [Crossref] [PubMed]
- Meng X, Shi Y, Xiang X, et al. Influence of miR-101 on proliferation of liver cancer cells through the MAPK/ERK signaling pathway. Oncol Lett 2020;19:1310-6. [PubMed]
- Hua FF, Liu SS, Zhu LH, et al. MiRNA-338-3p regulates cervical cancer cells proliferation by targeting MACC1 through MAPK signaling pathway. Eur Rev Med Pharmacol Sci 2017;21:5342-52. [PubMed]
(English Language Editor: A. Kassem)


