
B7-H3 promotes proliferation and migration of lung cancer cells by modulating PI3K/AKT pathway via ENO1 activity
Highlight box
Key findings
• B7-H3 can promote proliferation and migration of lung cancer cells by modulating PI3K/AKT pathway via ENO1 activity.
What is known and what is new?
• B7-H3 messenger RNA expression levels was up-regulated and correlated with patient prognosis in lung cancer.
• B7-H3 regulated ENO1 activity rather than changing its expression levels. B7-H3 can promote malignant phenotype of lung cancer cells by modulating PI3K/AKT pathway via ENO1 activity.
What is the implication, and what should change now?
• The results provide a vital theoretical basis for exploring the molecular mechanisms of lung cancer.
Introduction
Lung cancer, a malignant tumor with the highest mortality globally (1), can be divided into small cell lung cancer (SCLC) and non-SCLC (NSCLC) based on histological characteristics, and among which NSCLC accounts for more than 80% of the incidence population (2,3). Surgical treatment for patients with early lung cancer is the most effective therapeutic intervention, but most patients are already in advanced stage at initial diagnosis, with poor overall prognosis and low 5-year survival rates (4). Despite great advances in the development of molecular targeted therapy and immunotherapy, the prognosis of lung cancer has been bleak (5,6). Therefore, it is of great significance to comprehensively analyze the molecular mechanism of lung cancer and develop new immunotherapy targets for lung cancer treatment.
Recently, B7 superfamily of proteins has been highlighted for its remarkable therapeutic potential on several types of cancers (7). B7-H3, also known as CD276, was originally identified as a co-stimulatory molecule of the B7 family for T cell activation and IFN-γ production (8). Recent studies have shown that B7-H3 is involved in promoting carcinogenesis and metastasis in many kinds of malignant tumors, including glioblastoma (9), breast cancer (10), prostate cancer (11), ovarian cancer (12) and others. Emerging studies demonstrated that B7-H3 regulates a variety of signal transduction pathways related to tumor immune escape and metabolic dysregulation (13,14). Abnormal aerobic glycolysis and anabolic pathways are a major hallmark of cancer that can provide energy for tumorigenic processes (15,16). Picarda et al. highlighted that the loss of B7-H3 in adipocyte progenitors leads to impaired oxidative metabolic programs and increased lipid accumulation, affecting glycolysis and mitochondrial activity (17). Shi et al. demonstrated that treatment of cells with hexokinase 2 (HK2) inhibitors could reverse the increase in B7-H3-induced aerobic glycolysis (18). These studies support the involvement of B7-H3 in the metabolic disorders of cancer cells, but the mechanism is worth exploring.
ENO1, a rate-limiting enzyme associated with glycolysis, is overexpressed and activated in malignant tumor cells, contributing to accelerated glycolysis and tumor growth (19-21). Relevant studies have indicated that ENO1 primarily locates in the cytoplasm of cancer cells and regulates oncogenic signaling pathways like phosphorylation levels of PI3K/AKT pathways (22) to affect the proliferation and invasion of breast cancer cells and AMPK/mTOR pathway (23) to promote the self-renewal in lung cancer cells. Moreover, some circular RNAs (circRNAs) (24,25) and proteins (26,27) have also been reported to interact with ENO1 to regulate tumorigenesis, metastasis, and drug resistance, ENO1 has recently been considered as a tumor biomarker and clinical therapeutic target (28,29).
Previous study found that B7-H3 interacted with ENO1 to mediate the glycolysis process in cervical cancer cells (19). Through preliminary experiments and literature reviews, we found that B7-H3 and ENO1 have many similarities in lung cancer. However, the precise regulatory mechanism and their relationship between B7-H3 and ENO1 in tumor development are still need to be more deeply explored. In this study, we demonstrated that B7-H3 was up-regulated and correlated with poor prognosis in lung cancer. Functionally, B7-H3 knockdown could not only inhibit cell proliferation and migration but also affected ENO1 activity to further activate the PI3K/AKT pathway in lung cancer cells. Collectively, these results provide a vital theoretical basis for exploring the molecular mechanisms of lung cancer. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1537/rc).
Methods
B7-H3 expression-level analysis
The “Exploration-Gene DE” module of TIMER 2.0 (30) was used to assess B7-H3 expression in more than 30 tumors and nearby normal tissues. By using the “Expression DIY” module of GEPIA (31), B7-H3 expression between tumorous tissues of lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and normal surrounding tissues was analyzed. The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) (32) database was used to look into protein expression of B7-H3 in different patient groups. The Kaplan-Meier (KM) plotter (33) was used to analyze the correlation between B7-H3 and prognosis.
Cell culture
Four human lung cancer cell lines (A549, PC14, SBC5, and H446) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (Gibco, Carlsbad, NY, USA) supplemented with 10% fetal bovine serum (FBS; Every Green, Hangzhou, China) and 1% penicillin/streptomycin (Hyclone, Logan, UT, USA) under the conditions of 37 ℃ and 5% CO2.
Plasmid construction, small interfering RNAs (siRNAs) and cell transfection
The full-length complementary DNA (cDNA) of human B7-H3 was amplified from SBC5 cells and cloned into the expression vector pcDNA3.1. The constructed plasmid (pcDNA3.1-B7-H3) was verified by DNA sequencing and transfected into SBC5 cells with low expression of B7-H3 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Cell lines with stable expression of B7-H3 were selected using 800 µg/mL geneticin (Gibco) for follow-up experiments. Accordingly, specific siRNAs targeting human B7-H3 (siB7-H3), ENO1 (siENO1), and negative control (siNC) were designed and synthesized by Sangon Biotech (Shanghai, China). The transfection was performed in H446 cells with high expression of B7-H3 or ENO1 when cells confluency reached approximately 50–60%. Transfection efficiency was validated by real-time polymerase chain reaction (RT-PCR) and Western blot. The gene sequences targeting B7-H3 and ENO1 are described in Table S1.
Cell proliferation assay
Cell counting kit-8 (CCK-8) (KeyGEN Biotech, Nanjing, China) assay was used to examine cell proliferation following the manufacturer’s instructions. Cells transfected with overexpression plasmids or siRNAs were seeded into 96-well plates (2,000 cells/well in 100 µL of complete medium), respectively. At the indicated time points of 0, 24, 48, 72, and 96 hours, 10 µL of CCK-8 reagent was added to each well and incubated at 37 ℃ in the dark for 2 hours, then the absorbance in each well at 450 nm was measured by a multifunctional microplate reader (Biotek, Winooski, VT, USA). Cell growth curves were analyzed using GraphPad Prism software (GraphPad, San Diego, CA, USA).
Wound healing assay
Cell migration abilities can be observed and compared by the scratch test. Cells were seeded into six-well plates at a suitable density and incubated overnight to allow for cell attachment. When the cells grew to 85–90% confluence, wounded areas were created by gently scraping with a 200-mL pipette tip, and floating cells were removed. The cells were then cultured in serum-free medium for another 48 hours. The spacing of the gap was examined and photo imaged using an inverted microscope at 0 and 48 hours. Data were analyzed using GraphPad Prism.
Glutathione S-transferase (GST) pulldown assay
PCR primers for B7-H3 gene were obtained from Genepharma (Shanghai, China) according to the polyclonal restriction site of pGSTag prokaryotic expression vector and the CDS region of B7-H3 gene. PCR amplification products verified by sequencing were inserted into pGSTag vector to transform JM109 competent cells, and positive clones were selected to amplify and to extract plasmids. GST-B7-H3 chimeric protein was expressed in Escherichia coli BL21 (DE3) pLysS cells and induced by 0.1 mM isopropyl beta-D-1-thiogalactopyranoside (IPTG) for 12 hours at 24 ℃. The protein was purified by Sepharose 4B (Sigma, St. Louis, MO, USA) and identified by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The 50 µg purified GST-B7-H3 and GST-alone protein (as negative control) were separately incubated with Glutathione HiCap Magnetic Beads (Qiagen, Hilden, Germany) in a room temperature shaker at 4 ℃ for 6 hours, and then the beads mixtures were collected. H446 cell lysates were added to previous bead mixtures combined with GST-B7-H3 and GST protein, and incubated overnight on a 4 ℃ shaker, respectively. The next day, the precipitate was collected and washed for three times after removing the supernatant. The precipitated proteins were analyzed using specific primary antibody.
ENO1 enzyme activity assay
Enolase activity was determined using ENO1 human activity assay kit (Abcam, Cambridge, UK) according to the instructions provided by the manufacturer. Assays were performed using cell extracts from transfected cells, and quantifications were normalized to total protein contents determined by bicinchoninic acid (BCA). In brief, 10 µg cell lysates were added to a microplate coated with captured antibodies and incubated at room temperature for 2 hours. Subsequently, 200 µL of the reaction buffer (containing the substrate 2-phosphoglycerate, pyruvate kinase, and lactate dehydrogenase) was gently added to each well and immediately beginning to record the absorbance with elapsed time at 340 nm for 60 minutes. The consumption of NADH represented the enzymatic activity of ENO1 at the indicated time point.
ENO1 activity inhibition assay was also performed by AP-III-a4 (ENOblock), a novel small molecule inhibitor of α-enolase from MedChemExpress. Cells were treated with 1 µM ENOblock to inhibit the activity of ENO1. And then the inhibitory efficiency was detected as described above.
RT-PCR analysis
Total RNA was extracted from cells according to the protocol supplied with the TRIzol reagent (Takara, Kyoto, Japan). Isolated messenger RNA (mRNA) (1 µg) was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). Quantitative PCR was performed using SYBR Premix Ex Taq II (Takara) and amplification efficiencies were validated and normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Relative fold changes were analyzed using the 2−ΔΔCt method, and each sample was determined in triplicate. The mRNA-normalized data were expressed as relative gene mRNA in experimental group compared to control groups. The primer sequences used in the study are shown in Table S2.
Western blot analysis
Radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotech, Shanghai, China) containing protease inhibitor was used to isolate proteins from cells. An equal amount of protein (20 µg) was loaded with 12% SDS-PAGE before blotting onto polyvinylidene difluoride (PVDF) membrane (Merck, Darmstadt, Germany) under 15 V semi-dry for 40 minutes. The membrane was blocked with 5% skimmed milk in Tris-buffered saline with 0.05% Tween-20 for 2 hours at room temperature. Next, the membranes were probed with primary antibodies: anti-B7-H3 (Abcam, ab243883), anti-ENO1 (Abcam, ab155102), anti-PI3K p85α (Abcam, ab191606), anti-p-PI3K p85α (Abcam, ab182651), anti-AKT (Abcam, ab179463), anti-p-AKT (Abcam, ab38449) and anti-GAPDH (Abcam, ab8245) at 4 ℃ overnight, after washing, sheep anti-rabbit second antibody coupled to horseradish peroxidase (ZSGB-Bio, ZB-2301) was added for 1 hour at room temperature. Antibody-antigen complexes were then tested using enhanced chemiluminescence (ECL) (Bio-Rad Chemidoc MP, Singapore, Singapore). To assess protein expression, band intensity was quantified using IMAGEJ software.
Xenograft mouse models
Six-to-eight-week-old male C57BL/6 mice (n=20) were obtained from Beijing Vitonglihua Experimental Animal Technology Co., Ltd., and housed under specific-pathogen-free conditions. Approximately 2×106 Lewis lung cancer (LLC) cells transfected with shB7-H3 in 100 µL of phosphate-buffered saline (PBS) were implanted subcutaneously into the root of the left leg. At the end of the experiment, the mice were sacrificed by neck dislocation, and the tumor nodules were taken. The tumor volume (0.5 × length × width2) was measured every 4 days to monitor tumor growth. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. 20230832) granted by Air Force Medical University’s Institutional Animal Care and Use Committee, in compliance with Air Force Medical University’s guidelines for the care and use of animals.
Statistical analysis
The data were representative of at least three independent experiments as indicated. Survival curves were compared using KM analysis. Mann-Whitney U test was used for comparison between the two groups. Multiple comparisons were evaluated by repeated measures analysis of variance (ANOVA). All experimental data were analyzed using GraphPad Prism 8.0 software. P value less than 0.05 was considered statistically significant.
Results
B7-H3 was upregulated in lung cancer and correlated with tumor prognosis
To reveal the expression of B7-H3 in various cancers, we analyzed RNA-seq data of pan-cancer from the public database. As indicated in Figure 1A, the B7-H3 was highly expressed in lung cancer basing on the TIMER 2.0 database. Correspondingly, there was a rising trend in different types of tumors, including glioblastoma multiforme (GBM), kidney renal clear cell carcinoma (KIRC), and stomach adenocarcinoma (STAD), etc. Besides, we also confirmed significantly elevated expression of B7-H3 in lung cancer tissue (including LUAD, LUSC) compared with normal tissues, and it was closely correlated with clinical stage using the data from GEPIA2 and UALCAN database (Figure 1B-1D). Survival analyses showed that high levels of B7-H3 expression were markedly associated with poorer overall survival (OS) in lung cancer patient (Figure 1E).
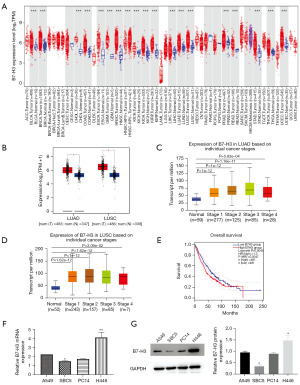
B7-H3 overexpression promoted cell proliferation and migration in lung cancer
The mRNA and protein levels of B7-H3 in four lung cancer cell lines (including A549, PC14, SBC5, and H446) were examined. The results showed that B7-H3 expression compared with A549 was lower in SBC5 and higher in H446 (Figure 1F,1G). Therefore, we selected these two different cell lines of lung cancer cells for follow-up tests.
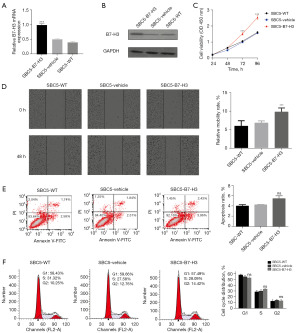
To investigate the effect of B7-H3 on malignant phenotype of lung cancer cells, we constructed the cell model that stably overexpressed B7-H3 and named as SBC5-B7-H3. The higher expression level of B7-H3 in SBC5-B7-H3 than that in SBC5-vehicle control cells was identified by quantitative RT-PCR (qRT-PCR) and immunoblotting (Figure 2A,2B). Then we used CCK-8 assay to analyze the proliferation of SBC5, and the result showed significant higher proliferation rate in SBC5-B7-H3 cells than that in control cells (Figure 2C). Moreover, wound healing assays showed that B7-H3 overexpression enhanced the migration ability of SBC5 cells (Figure 2D). However, there were no significant differences of cell cycle and cell apoptosis between SBC5-B7-B3 and SBC5-vehicle (Figure 2E,2F).
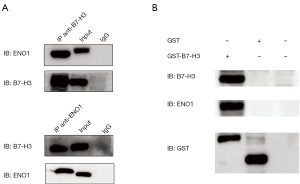
B7-H3 directly interacted with ENO1 in H446
To explore the underlying molecular mechanism of B7-H3 in the proliferation and migration of lung cancer cells, we carried out immunoprecipitation and GST pulldown experiment to confirm the ENO1 as B7-H3-binding enzyme proteins in lung cancer cells. We used B7H3 or ENO1 as a decoy protein to confirm the interaction between B7-H3 and ENO1 in H446 cell line by immunoprecipitation (Figure 3A). Additionally, we used the immobilized GST-B7-H3 and GST-alone (as a negative control) to incubate with cells lysates, respectively. Then, we observed specific bands around 47 kDa compared to negative controls (Figure 3B). The results showed that ENO1 bound to B7-H3 directly, suggesting that their interaction might be part of the mechanism by which B7-H3 promoted the proliferation and migration of lung cancer cells.
Silencing of B7-H3 or ENO1 could attenuate cell growth and migration in H446, affecting the PI3K/AKT signaling pathway
The function of B7-H3 and ENO1 was explored by knockdown gene expression using B7-H3 or ENO1 siRNA in H446 cells. The efficiency of knocking down was verified by qRT-PCR and immunoblotting (Figure 4A,4B). Since siB7-H3#2 and siENO1#1 were more effective in knocking down their respective target genes, these two siRNA were used for further experiment analysis. By CCK-8 and scratch assays, we found that B7-H3 or ENO1 knockdown could both inhibit cell proliferation and migration (Figure 4C,4D), respectively. Xenograft mouse models indicated that down-regulation of B7-H3 expression could significantly inhibit tumor growth in lung cancer (Figure 4E).
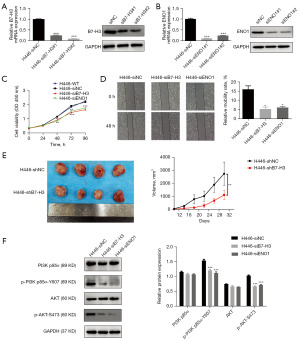
PI3K/AKT pathway plays a key role in lung cancer and is closely associated with prognosis and metastasis. To further understand whether the PI3K/AKT pathway is responsible for lung cancer progression, we also measured changes in PI3K/AKT signaling under conditions of B7-H3 or ENO1 knockdown. As a result, the phosphorylate levels of PI3K-p85α (Tyrosine 607) and AKT (Ser 473) were decreased by interference with B7H3 or ENO1 alone, whereas PI3K-p85α and AKT total protein levels were not altered (Figure 4F). These results suggested that PI3K/AKT may be a downstream common signaling pathway of B7H3 and ENO1 involved in the malignant phenotype of lung cancer cells.
B7-H3 regulated ENO1 activity rather than changing the expression levels
To assess the underlying correlations between B7-H3 and ENO1, we determined the influence of B7-H3 expression on ENO1 level changes by overexpression or knockdown of B7-H3 in SBC5 and H446 cells. However, we found no changes in either ENO1 transcription or protein levels after B7-H3 expression changes. Then, we tested the enzymatic activity changes of ENO1 in SBC5 and H446 cells, and found that overexpression of B7-H3 enhanced ENO1 activity, while inhibition of B7-H3 expression reduced its activity (Figure 5A,5B), suggesting that B7-H3 function on ENO1 was to regulate the enzymatic activity rather than to change the expression level.
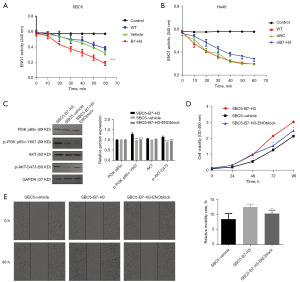
The effects of B7-H3 overexpression on lung cancer cells could be attenuated through blocking ENO1 activity
To further clarify the interaction mechanism between B7H3 and ENO1, we used AP-III-a4 to block ENO1 activity in SBC5-B7-H3 cells, then detected the changes of PI3K/AKT signaling pathway molecules and malignant phenotype. We found that the phosphorylation levels of PI3K-p85α (Tyrosine 607), and AKT (Ser 473) was expedited in SBC5-B7-H3 cells, while inhibition of ENO1 activity by AP-III-a4 reversed the above results (Figure 5C). Moreover, the proliferation and wound-healing speed of SBC5-B7-H3 cells were markedly increased compared with the control group, while significantly decreased after ENO1 activity was blocked (Figure 5D,5E). Thus, our data confirmed that interactions between B7-H3 and ENO1 promoted cancer cells progression mediated by upregulation of PI3K/AKT signaling pathway. All these results demonstrated that B7-H3 promotes proliferation and migration of lung cancer cells by modulating PI3K/AKT pathway via ENO1 activity.
Discussion
Despite recent improvements in diagnosis and treatment strategies, lung cancer patients still have a high mortality rate and poor prognosis. Recently, B7 family members have gained increasing attention due to their therapeutic effect on cancers, following the clinical success of inhibitory immune checkpoint blockade (CTLA-4, PD-1, and PD-L1) (34,35). B7-H3, as a novel member of B7 family, has emerged as the potential next promising target for cancer immunotherapy (36). However, since the receptor of B7-H3 has not been fully elucidated, its regulatory mechanism in promoting cancer progression is rarely reported.
B7-H3, also referred to as CD276, is a multifunctional protein that is abnormally expressed in diverse tumors and is involved in tumor genesis, metastasis, and immune evasion (37). Guo et al. studied the expression profiling of immunomodulatory-related genes in prostate cancers and showed remarkable differences in B7-H3 expression, which were linked to a dismal prognosis (38). Here, we utilized different public datasets to explore whether B7-H3 is involved in lung cancer progression. The results showed that B7-H3 was overexpressed in lung cancer tissues and was significantly correlated with individual cancer stage and survival probability, which just verified Guo et al.’s reports.
To elucidate the biological involvement of B7-H3 in lung cancer progression, we determined the expression of B7-H3 in four lung cancer cells, and selected appropriate cells to construct cell models. By cell experiments, we observed the promoting effect of B7-H3 overexpression on proliferation and migration ability of SBC5 cells. These data suggested that B7-H3 plays a carcinogenic role in lung cancer progression (21,22). However, there were no significant differences of cell cycle and cell apoptosis. The reasons may involve many factors, such as the activation of cell proliferation signaling pathway, the change of apoptosis regulation mechanism, and the expression of cell cycle regulation proteins. The interaction of these factors may result in increased cell proliferation while apoptosis and the cell cycle remain unchanged.
To further clarify the potential mechanism of B7-H3 in lung cancer, we performed GST pull-down and immunoprecipitation analysis in H446 cells and confirmed ENO1 as the B7-H3 partner molecule, which was also consistent with our previous research (19). Subsequently, we investigated the malignant phenotype of H446 cells with B7-H3 or ENO1 knockdown. By CCK-8 and scratch assays, we found that both B7-H3 and ENO1 knockdown could suppress the proliferation and migration of H446 cells. These results indicated that both B7-H3 and ENO1 are oncogenes and able to promote the progression of lung cancer.
ENO1 (α-enolase), expressed on the surface of the tumor cells, is implicated in the proliferation, invasion, and metastasis of cancer cells (29). In the cytoplasm, ENO1 is responsible for the transformation of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis, promoting the Warburg effect in tumor cells (39). More importantly, the researchers pointed out that some circRNA and proteins affected tumorigenesis and development by modulating ENO1 enzymatic activity (40-42). Considering that ENO1 is an important glycolytic enzyme, we determined the expression of ENO1 under condition of overexpression or knockdown expression of B7-H3. These results are also in accordance with previous reports that supporting an oncogenic role for ENO1 (20,22,23). However, we found no changes in either ENO1 mRNA or protein levels, suggesting that the interaction between B7-H3 and ENO1 may not be reflected in gene expression. In the meanwhile, inhibition of ENO1 activity could restore cell proliferation and migration by B7-H3 overexpression to normal levels. Zuo et al. used B7-H3 antibody to block the expression of B7-H3 in HeLa cells and found that the levels of c-Myc and LDHA genes associated with glycolysis were significantly reduced (19). It is suggested that B7-H3 may mediate glycolysis and promote tumor progression by regulating ENO1 activity induced by c-Myc and LDHA expression.
Accumulating evidence has suggested that PI3K/AKT is a common downstream signaling pathway of B7-H3 and ENO1, which is involved in tumorigenesis and metastasis (43,44). PI3K/AKT is a classical signal mediator during carcinogenesis and can promote cell proliferation, metabolism, and motility by activating downstream multiple effector molecules. We hypothesized whether PI3K/AKT contributes to the malignant behavior of ENO1-mediated progression in lung cancer cells. Then, we confirmed that B7-H3 overexpression significantly increased the protein levels of phosphorylated PI3K-p85α and AKT, while inhibition of ENO1 activity attenuated the effect of B7-H3 on the phosphorylate levels of these molecules. Conversely, silencing B7-H3 or ENO1 could attenuate the PI3K/AKT signaling pathway. These results verified our hypothesis, indicating that the ENO1-PI3K/AKT pathway was crucial in B7-H3-evoked enhancement of lung cancer progression.
Our study has several limitations. Previous studies revealed that B7-H3 was highly expressed in lung cancer, more clinical samples of lung cancer patients should be used to validate our results. Moreover, it is necessary to conduct more animal experiments to verify the mechanisms we proposed to explain our results.
Conclusions
Taken together, we illustrated the carcinogenetic effect of B7-H3 in lung cancer development. B7-H3 promote cell proliferation and migration by activating PI3K/AKT signal pathway via ENO1 activity. Therefore, our study provides new evidence that B7-H3 may be an option for the treatment of lung cancer.
Acknowledgments
Funding: This work was financially supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1537/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1537/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1537/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1537/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. 20230832) granted by Air Force Medical University’s Institutional Animal Care and Use Committee, in compliance with Air Force Medical University’s guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med 2021;27:1345-56. [Crossref] [PubMed]
- Liu SY, Bao H, Wang Q, et al. Genomic signatures define three subtypes of EGFR-mutant stage II-III non-small-cell lung cancer with distinct adjuvant therapy outcomes. Nat Commun 2021;12:6450. [Crossref] [PubMed]
- Yu X, Wang Z, Chen Y, et al. The Predictive Role of Immune Related Subgroup Classification in Immune Checkpoint Blockade Therapy for Lung Adenocarcinoma. Front Genet 2021;12:771830. [Crossref] [PubMed]
- Kontos F, Michelakos T, Kurokawa T, et al. B7-H3: An Attractive Target for Antibody-based Immunotherapy. Clin Cancer Res 2021;27:1227-35. [Crossref] [PubMed]
- Chapoval AI, Ni J, Lau JS, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol 2001;2:269-74. [Crossref] [PubMed]
- Wang MY, Qi B, Wang F, et al. PBK phosphorylates MSL1 to elicit epigenetic modulation of CD276 in nasopharyngeal carcinoma. Oncogenesis 2021;10:9. [Crossref] [PubMed]
- Shao L, Yu Q, Xia R, et al. B7-H3 on breast cancer cell MCF7 inhibits IFN-γ release from tumour-infiltrating T cells. Pathol Res Pract 2021;224:153461. [Crossref] [PubMed]
- Zhou Y, Zhang G, Zhang W, et al. B7-H3 Promotes Prostate Cancer Progression in Mice by Antagonizing Myeloid-Derived Suppressor Cell Apoptosis. Technol Cancer Res Treat 2020;19:1533033820971649. [Crossref] [PubMed]
- Miyamoto T, Murakami R, Hamanishi J, et al. B7-H3 Suppresses Antitumor Immunity via the CCL2-CCR2-M2 Macrophage Axis and Contributes to Ovarian Cancer Progression. Cancer Immunol Res 2022;10:56-69. [Crossref] [PubMed]
- Zhao B, Li H, Xia Y, et al. Immune checkpoint of B7-H3 in cancer: from immunology to clinical immunotherapy. J Hematol Oncol 2022;15:153. [Crossref] [PubMed]
- Getu AA, Tigabu A, Zhou M, et al. New frontiers in immune checkpoint B7-H3 (CD276) research and drug development. Mol Cancer 2023;22:43. [Crossref] [PubMed]
- Stine ZE, Schug ZT, Salvino JM, et al. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov 2022;21:141-62. [Crossref] [PubMed]
- You M, Xie Z, Zhang N, et al. Signaling pathways in cancer metabolism: mechanisms and therapeutic targets. Signal Transduct Target Ther 2023;8:196. [Crossref] [PubMed]
- Picarda E, Galbo PM Jr, Zong H, et al. The immune checkpoint B7-H3 (CD276) regulates adipocyte progenitor metabolism and obesity development. Sci Adv 2022;8:eabm7012. [Crossref] [PubMed]
- Shi T, Ma Y, Cao L, et al. B7-H3 promotes aerobic glycolysis and chemoresistance in colorectal cancer cells by regulating HK2. Cell Death Dis 2019;10:308. [Crossref] [PubMed]
- Zuo J, Wang B, Long M, et al. The type 1 transmembrane glycoprotein B7-H3 interacts with the glycolytic enzyme ENO1 to promote malignancy and glycolysis in HeLa cells. FEBS Lett 2018;592:2476-88. [Crossref] [PubMed]
- Chen JM, Chiu SC, Chen KC, et al. Enolase 1 differentially contributes to cell transformation in lung cancer but not in esophageal cancer. Oncol Lett 2020;19:3189-96. [Crossref] [PubMed]
- Jiang K, Dong C, Yin Z, et al. Exosome-derived ENO1 regulates integrin α6β4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell Death Dis 2020;11:972. [Crossref] [PubMed]
- Zang HY, Gong LG, Li SY, et al. Inhibition of α-enolase affects the biological activity of breast cancer cells by attenuating PI3K/Akt signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:249-57. [PubMed]
- Shu X, Cao KY, Liu HQ, et al. Alpha-enolase (ENO1), identified as an antigen to monoclonal antibody 12C7, promotes the self-renewal and malignant phenotype of lung cancer stem cells by AMPK/mTOR pathway. Stem Cell Res Ther 2021;12:119. [Crossref] [PubMed]
- Tang S, Nie X, Ruan J, et al. Circular RNA circNFKB1 promotes osteoarthritis progression through interacting with ENO1 and sustaining NF-κB signaling. Cell Death Dis 2022;13:695. [Crossref] [PubMed]
- Wang L, Li H, Qiao Q, et al. Circular RNA circSEMA5A promotes bladder cancer progression by upregulating ENO1 and SEMA5A expression. Aging (Albany NY) 2020;12:21674-86. [Crossref] [PubMed]
- Deng T, Shen P, Li A, et al. CCDC65 as a new potential tumor suppressor induced by metformin inhibits activation of AKT1 via ubiquitination of ENO1 in gastric cancer. Theranostics 2021;11:8112-28. [Crossref] [PubMed]
- Shen D, Deng Z, Liu W, et al. Melatonin inhibits bladder tumorigenesis by suppressing PPARγ/ENO1-mediated glycolysis. Cell Death Dis 2023;14:246. [Crossref] [PubMed]
- Almaguel FA, Sanchez TW, Ortiz-Hernandez GL, et al. Alpha-Enolase: Emerging Tumor-Associated Antigen, Cancer Biomarker, and Oncotherapeutic Target. Front Genet 2021;11:614726. [Crossref] [PubMed]
- Huang CK, Sun Y, Lv L, et al. ENO1 and Cancer. Mol Ther Oncolytics 2022;24:288-98. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 2021;19:4101-9. [Crossref] [PubMed]
- Zhang H, Dai Z, Wu W, et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer Res 2021;40:184. [Crossref] [PubMed]
- Zhao Y, Lee CK, Lin CH, et al. PD-L1:CD80 Cis-Heterodimer Triggers the Co-stimulatory Receptor CD28 While Repressing the Inhibitory PD-1 and CTLA-4 Pathways. Immunity 2019;51:1059-1073.e9. [Crossref] [PubMed]
- Yang S, Wei W, Zhao Q. B7-H3, a checkpoint molecule, as a target for cancer immunotherapy. Int J Biol Sci 2020;16:1767-73. [Crossref] [PubMed]
- Mortezaee K. B7-H3 immunoregulatory roles in cancer. Biomed Pharmacother 2023;163:114890. [Crossref] [PubMed]
- Guo C, Figueiredo I, Gurel B, et al. B7-H3 as a Therapeutic Target in Advanced Prostate Cancer. Eur Urol 2023;83:224-38. [Crossref] [PubMed]
- Cancemi P, Buttacavoli M, Roz E, et al. Expression of Alpha-Enolase (ENO1), Myc Promoter-Binding Protein-1 (MBP-1) and Matrix Metalloproteinases (MMP-2 and MMP-9) Reflect the Nature and Aggressiveness of Breast Tumors. Int J Mol Sci 2019;20:3952. [Crossref] [PubMed]
- Li J, Hu ZQ, Yu SY, et al. CircRPN2 Inhibits Aerobic Glycolysis and Metastasis in Hepatocellular Carcinoma. Cancer Res 2022;82:1055-69. [Crossref] [PubMed]
- Li HJ, Ke FY, Lin CC, et al. ENO1 Promotes Lung Cancer Metastasis via HGFR and WNT Signaling-Driven Epithelial-to-Mesenchymal Transition. Cancer Res 2021;81:4094-109. [Crossref] [PubMed]
- Zhang G, Zhao X, Liu W. NEDD4L inhibits glycolysis and proliferation of cancer cells in oral squamous cell carcinoma by inducing ENO1 ubiquitination and degradation. Cancer Biol Ther 2022;23:243-53. [Crossref] [PubMed]
- Liao H, Ding M, Zhou N, et al. B7-H3 promotes the epithelial-mesenchymal transition of NSCLC by targeting SIRT1 through the PI3K/AKT pathway. Mol Med Rep 2022;25:79. [Crossref] [PubMed]
- Li Y, Li Y, Luo J, et al. FAM126A interacted with ENO1 mediates proliferation and metastasis in pancreatic cancer via PI3K/AKT signaling pathway. Cell Death Discov 2022;8:248. [Crossref] [PubMed]


