
WTAP-mediated abnormal m6A modification promotes cancer progression by remodeling the tumor microenvironment: bibliometric and database analyses
Highlight box
Key findings
• Wilms’ tumor 1 associated protein (WTAP)-mediated abnormal m6A modification promotes cancer progression by remodeling the tumor microenvironment.
What is known and what is new?
• The role of m6A methylation in tumor microenvironment (TME) is not negligible, and m6A methylation in tumor cells will affect the infiltration, activation, and efficacy of immune cells in TME.
• The abnormal modification of m6A mediated by WTAP can affect cancer occurrence, development, and metastasis by reshaping TME.
What is the implication, and what should change now?
• This study would provide helpful references for scholars focusing on WTAP and provide new insights for WTAP as a prognostic evaluation and immunotherapy for tumors in the future.
Introduction
Wilms’ tumor 1 associated protein (WTAP) is a protein that interacts with the Wilms’ tumor 1 (WT1) protein identified in yeast two-hybrid screening, and its gene is located on human chromosome 6q25-27 (1). m6A modification is a process in which methyltransferase catalyzes the methylation of adenine at the N6 position, which is the most abundant epigenetic modification in eukaryotic mRNA (2) and is involved in many vital processes such as the growth and development of mammals and the occurrence and development of diseases (3). Bokar et al. discovered that the methyltransferase complex could catalyze the formation of m6A in 1994 (4), and the methyltransferase METTL3 was found in 1997 (5). In 2014, Ping et al. first identified WTAP as a critical component of the mammalian m6A methyltransferase complex (6). Since then, the research field of methyltransferase and m6A modification has been opened and has attracted the interest of many researchers. As an essential regulatory subunit for methyltransferase activity, WTAP plays a crucial role in the occurrence and development of many diseases by regulating m6A modification (6). In particular, WTAP expression is upregulated in most tumor cells and is associated with poor prognosis. Mechanistically, WTAP mainly affects the expression level of m6A on the mRNA of target genes, thus regulating the expression of target genes and activating or inhibiting related pathways to affect tumor progression (7). It is worth noting that WTAP-mediated changes in m6A expression levels showed different regulatory results in different target genes, the molecular mechanism of which requires further study. In addition, WTAP also plays an alternative splicing function to regulate the gene transcription process, which directly or indirectly affects embryonic development (8). The current studies on the mechanism of WTAP-mediated m6A in different tumors and its biological function, as well as the clinical protocols of WTAP as a potential therapeutic target are of great significance. In recent years, the research interest in m6A modification has remained high, and the number of relevant papers continues to surge yearly. As a critical protein involved in m6A modification, WTAP has also received extensive attention from scholars around the world. Comprehensive bibliometric and database analyses of the WTAP research area are needed thereby.
m6A is the most abundant RNA modification in eukaryotes, and in recent years, more and more evidence has proved that m6A modification plays an important role in the pathogenesis of tumors such as glioblastoma, hepatocellular carcinoma, renal cell carcinoma, breast cancer, and acute myeloid leukemia, etc. (9-11). m6A promotes or suppresses tumor cells mainly by regulating the expression of mRNAs of relevant oncogenes or tumor suppressor genes, and the level of expression often directly determines the pathology of tumors. m6A methylation modification is a double-edged sword. The level of m6A methylation modification is a double-edged sword, and over-modification or reduction of the modification level may lead to tumor development and progression (12,13). Therefore, the study of m6A-regulated genes in different cancers and the identification of key m6A target genes are of great significance to the understanding of cancer pathogenesis and the development of individualized therapeutic plans.
The tumor microenvironment (TME) plays a crucial role in tumor development. It comprises cellular components and an extracellular matrix, in which infiltrating immune cells account for a large proportion (14). Commonly, immune cells recognize and eliminate abnormal tumor cells (15). However, tumor cells can inhibit the anti-tumor effect of the human immune system in various ways to survive and grow (16). The infiltrating immune cells and stromal cells in the TME are associated with angiogenesis, chemotherapy resistance, immunosuppression, and tumor cell migration (17,18). Previous studies have shown that the role of m6A methylation in TME is not negligible, and m6A methylation in tumor cells will affect the infiltration, activation, and efficacy of immune cells in TME, and promote the formation of TME and cellular adaptation (19). TME also plays a crucial role in the complex regulatory network of m6A modification. TME can affect the expression of related regulatory factors of m6A modification through hypoxia-inducible factors (HIFs) and other factors, thereby changing the abundance of m6A. In short, m6A and TME are mutually induced and regulated in tumor occurrence and development, both complementing each other and supplementing mutually (20).
Bibliometrics is an interdisciplinary subject that uses mathematical and statistical methods to quantitatively analyze and study the distribution structure, quantitative relationship, and development law of literature with the literature system and bibliometric characteristics as the research object (21). Bibliometric techniques can offer a convenient way to visibly measure researchers’ efforts in the investigation of a specific field. With the help of VOSviewer1.6.18 and Citespace5.8R3, we conducted bibliometric analysis on WTAP research, quickly obtained important information from massive data, constructed a visual knowledge map, summarized the research hotspots and development trends in this research field, and looked for new topics and directions (22,23). We also extracted the data of WTAP expression in pan-cancers from the online databases, and analyzed its relationship with survival prognosis of the patients.
Methods
Data collection
The Web of Science Core Collection (WoSCC) database is considered the most influential database, and we chose it because it can provide comprehensive information bibliometric software needs (24). We conducted data retrieval in the WoSCC database. The retrieval time was from the beginning of the database to 6 June 2022. WTAP and its MESH words were used as search terms, the literature types were Article and Review, and the language was limited to English. The search results were deduplicated, and non-representative articles such as conference calls, news, and reviews were deleted. The downloadable search results record reads “Full Record and Cited References”. By 6 June 2022, 691 articles were finally selected from the database, including 636 articles and 55 reviews. In addition, we obtained the gene data of various tumors and normal tissues from TCGA and GTEx databases, and performed pan-cancer analysis and immunoassay for WTAP using the Sangerbox 3.0 online tool. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data analysis and visualization
There is currently much auxiliary software for bibliometric analysis, including VOSviewer, CiteSpace, SCI2, NetDraw, HistCite, etc. (25-28). Comparing the characteristics and advantages of each software, we used VOSviewer and CiteSpace to conduct bibliometric analysis and draw the knowledge map (27,29,30). We imported the “download_*.txt” file into CiteSpace 5.8.R3 and selected “Data” to remove duplicated studies. The time span was set as 1999–2022.06 and years per slice. Keywords were merged with the same meaning in the exported data and meaningless keywords were removed. The author keywords were used in VOSviewer1.6.18 for keyword co-occurrence analysis, and the counting methods were all fractional counts (31). Thresholds (T) of items were set based on different situations, which were marked in corresponding tables and figures. Microsoft Office Excel 2019 is used to draw a three-line chart of keywords and co-cited references. Besides, the 2020 journal impact factor (IF) and JCR were obtained from the Web of Science.
Results
Annual growth trend of the papers on WTAP research
A total of 691 articles were included, including 636 articles and 55 reviews. As can be seen from Figure 1A, since 1999, the annual publication volume and citation frequency of WTAP-related research in journals showed a steady upward trend, especially in the four years from 2017 to 2021, which almost showed a fold surge. Figure 1B is the annual publication volume trend chart of each country drawn on the bibliometrics online analysis platform, which can more intuitively show the differences in publication volume of different countries at different time points. Colors in the figure represent different countries, among which China, represented by blue, is the country with the most significant number of publications and the fastest trend growth in recent years.

Countries/regions and institutions in WTAP research
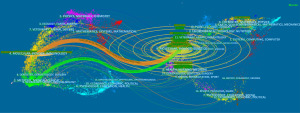
The top five countries in the number of WTAP research papers were China (n=288), the United States (n=191), Japan (n=58), Germany (n=55) and the United Kingdom (n=40). China had the largest number of documents, accounting for about 41.68%, but the betweenness centrality is only 0.05, while that of the United States (0.4) is the highest among all countries. Betweenness centrality (≥0.10) is usually regarded as the important turning point that may lead to transformative discoveries and acts as a bridge (32). Figure 2A contains 49 nodes and 164 links, with a density of 0.1395, indicating active collaborations among different countries/regions.

The top 10 institutions with the most significant number of articles include 8 from China, 1 from Japan and 1 from the United States. Among them, China Medical University is the institution with the most significant number of articles (n=19), accounting for about 2.75% of the total, followed by Zhejiang University in China (n=18), University of the Chinese Academy of Sciences (n=16), Zhengzhou University (n=15), and Osaka University in Japan (n=14). Close cooperation between different institutions can be observed (Figure 2B).
Journals and co-cited journals publishing WTAP research
VOSviewer was used for visual analysis of journals and co-cited journals which published the WTAP research. Among the 371 academic journals, Frontiers in Cell and Developmental Biology was the journal with the most significant number of articles (n=11), followed by Journal of the American Society of Neurology (n=10), Frontiers in Oncology (n=9), Journal of Biological Chemistry (n=9), and Human Molecular Genetics (n=9). Five of the top 10 journals were in Q1, the IFs of 9 journals were above 5, and most of them were from the United States (n=4) and the United Kingdom (n=3). These journals were the most influential core journals in the field of WTAP research.
Among the 2,933 co-cited journals, 10 journals had citations over 496. Nature had the most co-citations (n=1,256, 5.55%), followed by Cell, Journal of Biological Chemistry, and Proc Natl Acad Sci USA. Among the top 10 co-cited Journals, 9 were distributed in Q1, and IFs of 8 journals were more than 10, all from the United States (n=7) and the United Kingdom (n=3).
The dual-map overlay of journals stands for the topic distribution of academic journals (33), and that of WTAP papers is shown in Figure 3. The citing journals were located on the left, the cited journals were on the right, and the colored paths indicated the citation relationships. The diagram mainly includes an orange and green citation path. The orange course suggests that the articles published in Molecular/Biology/Genetics were cited primarily by the articles published in Molecular/Biology/Immunology. The green path indicates that articles published in Molecular/Biology/Genetics were mainly cited by papers published in Medicine/Medical/Clinical.
Authors in WTAP research
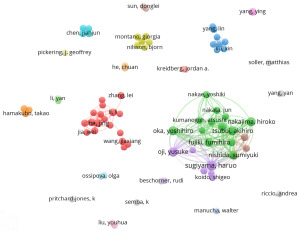
Haruo Sugiyama was the author with the most significant number of papers in the WTAP research field (n=17), followed by Yusuke Oji (n=12) and Yoshihiro Oka (n=11). Although the number of He Chuan publications was only five, the total citation times were as high as 2020. A total of 4,771 authors participated in the study. The author cooperation network diagram is constructed with the authors who have published at least three papers (Figure 4), the same color represents the same cluster, and the cooperation among authors in the same collection is relatively close; some authors also have cooperative relationships in different groups, such as He Jing and Wang Jia Xiang, Nakajima Hiroko and Kyodo Shigeo.
Keyword co-occurrence, clusters, and evolution
Keywords can not only express the most core and essential part of an academic paper clearly and intuitively, but also have the functions of indexing, quick reading, and highlighting the key points. VOSviewer was used to present the keyword co-occurrence and cluster analysis (Figure 5), replace and merge the synonyms before visualization, such as incorporating “N6-methyladenosine” and “m6A” into “m6A”, and then deleting the keywords without obvious research significance, such as “disease” and “mouse”. The top 20 high-frequency keyword tables were m6A, WT1, Wilms tumor, prognosis, immunohistochemistry, AML, immunotherapy, METTL3, apoptosis, podocytes, biomarkers, cancer, proliferation, ovarian cancer, cancer vaccine, FTO, metastasis, breast cancer, METTL14, and epigenetics. High-frequency keywords show that WTAP research mainly focuses on m6A modification and tumors. Cluster analysis can display the knowledge structure of the research field. The visualization map is clustered according to the co-occurrence link strength (Figure 5). The keywords of the red cluster are m6A, METTL3, METTL14, ALKBH5, FTO, apoptosis, cell cycle, inflammatory reaction, oxidative stress, DNA methylation, transcription, epigenetic transcriptome, p53, oncogene, proliferation, invasion, and metastasis, and the research directions involved in these keywords are m6A modification and its regulatory mechanism, and the biological characteristics of tumors. The keywords of the green cluster include WT1, nephroblastoma, cancer, tumor vaccine, immunotherapy, cisplatin, genechip technology, experimental embryology, dendritic cells, leukemia, breast cancer, renal cell carcinoma, which are closely related to tumor and its treatment. The blue cluster has minor keywords, including prognosis, biomarkers, bioinformatics, tumorigenesis, TCGA, immunohistochemistry, lung cancer, liver cancer, and ovarian cancer, which are related to tumor markers, and prognosis. It can be seen from the chart that m6A is the most critical term, with a total of 109 occurrences (Table 1).
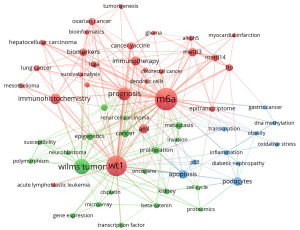
Table 1
| Rank | Term | Count |
|---|---|---|
| 1 | m6A | 109 |
| 2 | WT1 | 91 |
| 3 | Wilms tumor | 52 |
| 4 | Prognosis | 43 |
| 5 | Immunohistochemistry | 24 |
| 6 | AML | 18 |
| 6 | Immunotherapy | 18 |
| 8 | METTL3 | 17 |
| 8 | Apoptosis | 17 |
| 10 | Podocytes | 16 |
| 11 | Biomarkers | 14 |
| 11 | Cancer | 14 |
| 13 | Proliferation | 12 |
| 13 | Ovarian cancer | 12 |
| 13 | Cancer vaccine | 12 |
| 13 | FTO | 12 |
| 17 | Metastasis | 12 |
| 17 | Breast cancer | 11 |
| 19 | METTL14 | 11 |
| 20 | Epigenetics | 10 |
WTAP, Wilms’ tumor 1 associated protein; AML, acute myeloid leukemia; FTO, fat mass and obesity-associated protein.
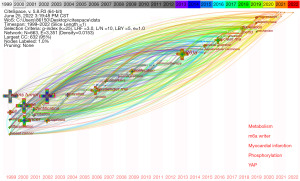
The timezone view can locate the time point when new keywords first appear and can quickly understand the development context of relevant fields, predict the development direction, and find research hotspots (26). In our analysis on WTAP research, the threshold value is set to 15, excluding keywords that only appear once or twice. Designed with CiteSpace software, the keyword’s timezone view is shown in Figure 6. New high-frequency keywords in the last three years are displayed in different colors at the bottom right of the timezone view. In the last three years, the top five keywords were metabolism, m6a writer, myocardial infarction, phosphorylation, and Yes-associated protein (YAP), showing the new direction of WTAP research.
Co-cited reference and reference burst
In 1973, the American information scientist Small first proposed the concept of co-cited reference as a research method to measure the degree of relationship between documents. Through the co-cited network, we can explore the development and evolution of a specific field. The VOSviewer was used to detect the co-cited references (Table 2). The cited frequency of the top 10 co-cited references was at least 43 times, and the most mentioned was the article, “Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase”, published by Ping in 2014 (6). This study identified for the first time that WTAP and METTL14 are essential components of the mammalian m6A methyltransferase complex.
Table 2
| Rank | Reference | Journal | Citation |
|---|---|---|---|
| 1 | Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase | Cell Research | 125 |
| 2 | Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq | Nature | 98 |
| 3 | A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation | Nature Chemical biology | 98 |
| 4 | ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility | Molecular Cell | 87 |
| 5 | N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO | Nature Chemical Biology | 86 |
| 6 | Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 50 sites | Cell Reports | 63 |
| 7 | Identification of WTAP, a novel Wilms’ tumour 1-associating protein | Human Molecular Genetics | 62 |
| 8 | m(6)A RNA methylation promotes XIST-mediated transcriptional repression | Nature | 47 |
| 9 | Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle | The Journal of Biology Chemistry | 44 |
| 10 | WTAP is a novel oncogenic protein in acute myeloid leukemia | Leukemia | 43 |
WTAP, Wilms’ tumor 1 associated protein.
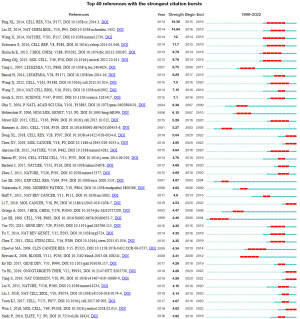
References with citation bursts are defined as those that are cited frequently over a while (34). The burst duration is set to 2 years on the CiteSpaces, γ is set to 1, and a total of 64 documents meeting the requirements are detected; we select the top 40 papers with burst intensity and visualize them (Figure 7). According to this figure, there are four articles with a burst passion of more than 10, of which the highest burst intensity is 16.36, which is the article published by Ping on Cell Research in 2014, and also the paper cited the most times, indicating its milestone significance in the WTAP field. Up to now, there were still six references in a continuous burst, which can be used to analyze the frontier dynamics and development trends of this research field.
Pan-cancer analysis and immunoassay
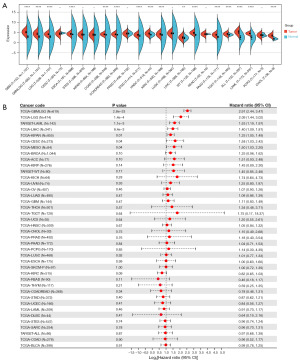
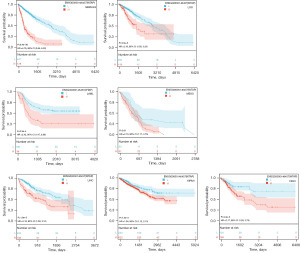
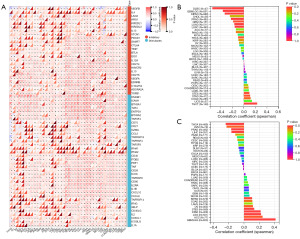
We performed WTAP gene expression differential analysis and prognostic analysis of 23 tumors from the TCGA and GTEx databases using the Sangerbox3.0 online tool. The gene differential expression analysis results showed that WTAP was highly expressed in 16 tumors, including GBM, GBMLGG, LGG, ESCA, COAD, COADREAD, STAD, HNSC, KIRC, LIHC, WT, PAAD, TGCT, ALL, LAML, and CHOL (Figure 8A, the whole tumor names are listed in Table S1). The relationship between WTAP expression and overall survival of cancer patients was analyzed for 40 tumors (Figure 8B). Predictive analysis showed that the high expression of WTAP in GBMLGG, LGG, LAML, MESO, LIHC, KIPAN and CESC was an essential factor affecting the poor prognosis of patients (Figure 9). Clinical staging and gene expression analysis suggested that WTAP expression in KIPAN and BLCA tumors affected tumor invasion and metastasis (KIPAN: P=0.006; BLCA: P=0.04).
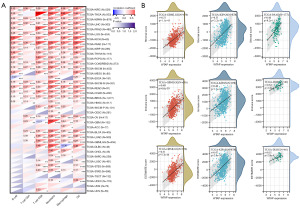
A standardized pan-cancer dataset, TCGA TARGET GTEx (PANCAN, N=19,131, G=60,499), was downloaded from the UCSC database for immune correlation analysis. The Timer method of R software package IOBR was used to evaluate the infiltration scores of B cell, T cell CD4, T cell CD8, neutrophil, macrophage, and DC of each patient tumor. The results showed that the expression of WTAP was correlated with six types of immune cells in most tumors (Figure 10A). The stromal cells, immune cells, and ESTIMATE scores of WTAP in each cancer were calculated using the R software package ESTIMATE and visualized using scatter plots. The results showed that WTAP had the strongest positive correlation with the stromal cell scores of GBMLGG, KIPAN, and PAAD, with the immune cell scores of GBMLGG, KIPAN, and DLBC, and with the ESTIMATE scores of GBMLGG, KIPAN, and DLBC (Figure 10B).
Immune checkpoint gene analysis showed that WTAP expression levels significantly correlated with these 40 checkpoint genes in numerous tumors (Figure 11A). Tumor mutation burden (TMB) and microsatellite instability (MSI) are emerging predictive markers for tumor immunotherapy. This study found that in GBMLGG, LGG, STES, KIPAN, STAD, OV, and ACC, the expression of WTAP was positively correlated with TMB, while in PRAD, LIHC, THCA, and UVM, the expression of WTAP was negatively correlated with TMB. It strongly correlates with GBMLGG (R=0.41) (Figure 11B). The expression of WTAP was positively correlated with MSI in COAD, STES, STAD, and TGCT. In GBM, GBMLGG, LUAD, KIPAN, PRAD, LUSC, THCA, and DLBC, the expression of WTAP was negatively correlated with MSI, and the strongest correlation was found with DLBC (R=0.54) (Figure 11C).
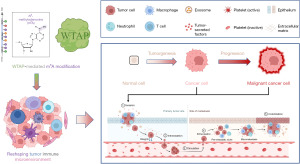
Summary of the mechanism of WTAP in cancer progression
The mechanism of WTAP in cancer progression is summarized in Figure 12 and will be elaborated in the Discussion. Previous studies have shown that m6A methylation in tumor cells will affect the infiltration, activation, and efficacy of immune cells in TME, and promote the formation of TME (19). WTAP regulates N6A methylation modification and participates in the occurrence and development of tumor via the effects of m6A. Our analysis revealed that the expression of WTAP was significantly correlated with immune infiltration in most tumors, and WTAP was most strongly associated with stromal cell scores and immune cell scores. These results suggest that WTAP is highly involved in tumor immune microenvironment remodeling.
Discussion
General information
In this study, CiteSpace and VOSviewer were used to conduct bibliometric analysis on the documents collected in the WOSCC database and to manually mine and sort out the node information of the visual atlas. Through the atlas, the cooperative network relationships among the countries, institutions, authors, journals, keywords, and references included in the literature are intuitively displayed, the research status in the field of WTAP is summarized and analyzed, and research hotspots and emerging topics are explored, looking forward to the future development trend. We collected and collated the relevant literature on WTAP research in the core collection database of Web of Science from January 1999 to June 2022. A total of 691 papers were included; these papers came from 49 countries, 1,071 institutions, and 4,771 authors and were published in 371 academic journals. The annual publication volume is an important indicator showing the development trend of the research field (35). Since 1999, the number of publications on WTAP-related research has gradually increased, attracting more and more attention from researchers.
There are significant differences in WTAP research among different countries and institutions. The top three countries with the most important number of papers were China, the United States, and Japan. The United States is not only the main cooperation center in this field but also has made milestone contributions to research progress and breakthroughs. Germany, Britain, and France are also significant to the development of WTAP. Most of the top ten institutions with the most important number of articles were from China. China Medical University was the largest institution with the most significant number of pieces, followed by Zhejiang University and the University of the Chinese Academy of Sciences. Osaka University in Japan also ranks fifth in the number of articles, but the cooperation between Osaka University and other institutions is not close. Among the top 100 highly cited papers, China is still the country with the most significant number of articles and has frequent academic exchanges with the United States, Britain, and other countries. Through the visual knowledge map, we find that there are active cooperation and exchange relations between different countries and different institutions. The research in the field of WTAP has attracted extensive attention worldwide.
The analysis of journals and co-cited journals showed that Frontiers in Cell and Developmental Biology was the journal with the most significant number of articles, and Nature was the co-cited journal with the most important number of citations. Among the top 100 highly cited papers, Frontiers in Oncology was the journal with the most significant number of articles, and Nature was still the journal with the most important number of citations. WTAP research was mainly concentrated in molecular biology, consistent with the results shown in the journal overlay view. The journal double view overlay represents the theme distribution of academic journals. The overlay map drawn by CiteSpace mainly has two connecting paths. The orange path indicates that the articles published in the journal Molecular/Biology/Genetics were particularly cited by those published in the journal Molecular/Biology/Immunology. The green path represents that the literature published in Molecular/Biology/Genetics were mainly cited by the articles published in Medicine/Medical/Clinical. Currently, WTAP-related research focuses on basic medical research, but it is still exploring and developing in clinical medicine and pharmaceutical research. In the future, WTAP is expected to become a piece of good news for cancer patients.
The number of published articles can analyze the core authors who greatly influence this research field. Haruo Sugiyama was the author with the most published reports, followed by Yusuke Oji and Yoshihiro, all leading figures related to WTAP research. Researchers in this field have active collaborative relationships within or between institutions, especially among high-impact authors. Through bibliometric analysis, we can get relevant information about potential collaborators and influential research groups, which can provide direction and guidance for the subsequent research of scholars.
Knowledge base
The knowledge base is a collection of commonly cited references cited by corresponding research groups (30,36), which is not entirely equivalent to highly cited references. The knowledge base of the WTAP research field was evaluated by analyzing the top 10 co-cited references. In this bibliometric analysis, the ten co-cited references were as follows: (I) in 2014, Cell Research published the most co-cited study authored by Ping and other scholars outstanding in WTAP research. In this paper, it was first confirmed that WTAP and METTL14 are essential components of the mammalian m6A methyltransferase complex; WTAP is a regulatory subunit necessary for m6A methyltransferase activity and plays a crucial role in the transcriptional regulation of RNA metabolism. (II) The second co-cited publication in Nature in 2012 by Dominissini et al. The research results showed that the RNA modification of m6A plays a vital role in regulating gene expression. Silencing methyltransferase will significantly affect gene expression and alternative splicing, thus affecting the TP53 signaling pathway and cell apoptosis (37). (III) Liu et al. published the third co-cited study in Nature Chemical Biology in 2014, which proposed that WTAP interacts with METTL3 and METTL14 and co-localizes with METTL3-METTL14 heterodimers in the nucleus to participate in m6A methylation modification. This paper demonstrated that WTAP alone had no methyltransferase activity in vitro. However, knockdown of WTAP could significantly reduce m6A levels (38). (IV) Zheng’s work published in Molecular Cell in 2013 showed that ALKBH5, as another mammalian RNA demethylase, can remove m6A modification from mRNA both in vivo and in vitro, thus affecting RNA metabolism and gene expression (39). (V) In 2011, Jia et al. published the fifth co-cited paper in Nature Chemical Biology, which shows that FTO exhibits efficient oxidative demethylation activity in vitro. It was also proved that m6A, as the physiological substrate of FTO, may affect the processing of pre-mRNA or nuclear RNAs (40). The fourth and fifth articles are all about the study of m6A demethylase (ALKBH5, FTO), which acts as the “eraser” of m6A RNA and the “writer” of m6A RNA (METTL3, METTL14, WTAP) to demethylate and methylate RNA respectively, and maintain the dynamic balance between deposition and clearance of m6A modification, which is essential for normal biological processes and development. (VI) In 2014, Schwartz et al. published the sixth co-cited paper on Cell Reports, the novel components of methylated transferase complex (WTAP, METTL14, and KIAA1429) were identified by proteomic screening and experimentally verified to be essential for m6A methylation. This study also showed that WTAP-dependent methylation is closely related to mRNA stability (41). (VII) The seventh reference, published in 2000 in Human Molecular Genetics by Little et al., was the first to identify a nuclear protein that interacts with the WT1 gene, in a yeast two-hybrid screen, known as WTAP (1). (VIII) The eighth co-cited publication was published in Nature in 2016 by Patil et al., these studies have shown that m6A methylation can promote an extended non-coding RNA XIST mediated gene silencing function, and WTAP can also regulate this function by promoting m6A methylation (42). (IX) Horiuchi et al. published the ninth paper in The Journal of Biology Chemistry in 2013. This study found that the WTAP complex regulates the alternative splicing of pre-mRNA by promoting the production of truncated isomers, which leads to a change in WTAP protein expression. The experiment verified that the consumption of WTAP complex components would lead to the arrest of the cell cycle, indicating that WTAP plays an essential role in the regulation of the cell cycle (43). (X) The last co-cited reference was published by Bansal et al. in 2014, which verified that WTAP, as a novel oncogenic protein in AML, plays an oncogenic role in AML by promoting the abnormal proliferation of bone marrow cells and blocking the terminal differentiation of bone marrow cells (44). The top 10 co-cited references focused on m6A modification, mainly including the mechanism of methyltransferase (WTAP, METTL3, METTL14) and demethylase (ALKBH5, FTO), and the correlation of WTAP regulating gene expression through m6A methylation modification, and the influence of abnormal expression of WTAP on the occurrence, development, and treatment of some diseases.
Research hotspots and emerging topics
In bibliometrics, keywords can reflect the research frontiers and hotspots in an academic field (45). The timezone view can show the evolution process of research hotspots more intuitively and even predict the research trend in the future (46). The most frequent keyword was m6A, followed by WT1, Wilms tumor, prognosis, etc. WTAP research is in the stage of rapid development, and the emerging keywords mainly include m6A modification, methyltransferase, P53, RNA translation, ovarian cancer, demethylation, phosphorylation, etc., representing the hot spot in recent years. References with high burst intensity can also characterize an emerging topic in an academic field (35,47). Half of the connections with top 10 intensive burst references coincide with the top 10 co-cited references, which indicates that concerns with higher burst intensity are more likely to become co-cited references with higher cited frequency. This consistency also reflects the influence of papers with high burst intensity in this research field. Six articles of top 40 authorities with the most vigorous citation burst are still in business, and they are following. (I) Review by Deng et al. published in Cell Research in 2018, which summarized the m6A “writers” (METTL3, METTL14, WTAP), “Eraser” (FTO, ALKBH5); “reader” (IGF2BP) in different tumors, and the intervention measures against cancer caused by dysregulation of m6A modification (48). (II) An article in Nature Communications by Yang et al. showed that FTO, as an m6A demethylase, plays a crucial role in promoting melanoma development and resistance to anti-PD-1 immunotherapy, the combination of FTO inhibition and anti-PD-1 blockade may reduce the resistance of melanoma to immunotherapy (49). (III) The article summarizes recent advances in the biological functions of m6A methylation modifications in cancer and discusses potential therapeutic strategies (50). (IV) An article published by Li et al. on Molecular Cancer in 2019 demonstrated that METTL3 promotes the development of colorectal cancer through an m6A-IGF2BP2-dependent regulatory mechanism (51). Relevant references also pointed out that WTAP, as an oncogene, promotes the progression of colorectal cancer through the WTAP/WT1/TBL1 axis in the Wnt signaling pathway (52). (V) The study published on Molecular Cell in 2019 by Wen et al. found that Zc3h13, an essential regulatory protein of m6A, can interact with WTAP, Virilizer and Hakai proteins to form a regulatory complex and anchor it in the nucleus to promote m6A methylation repair and regulate the self-renewal of mESCs (mouse embryonic stem cells) (53). (VI) The study showed that WTAP is highly expressed in high-grade serous ovarian cancer (HGSOC) and plays its function by promoting the proliferation and migration of ovarian cancer cell lines and inhibiting cell apoptosis. After preliminary experiments, it was speculated that the mechanism of WTAP may be related to MAPK and AKT pathways (54); the WTAP gene is localized on human chromosome 6q25-27 (1), which is also a chromosomal region of ovarian cancer (55), demonstrating the potential correlation between WTAP and HGSOC. In this study, the reference burst and keyword co-occurrence analysis show that the research topics and hotspots in the field of WTAP are concentrated in the following three directions.
The first is that WTAP regulates m6A methylation modification and relies on m6A to participate in the occurrence and development of diseases. WTAP is a methyltransferase without a catalytic domain and has no catalytic capacity for RNA alone, but it can stabilize the core complex and promote the localization of the METTL3-METTL14 complex in nucleophiles rich in mRNA splice genes and enucleation transporters (6). At the same time, the down-regulation of WTAP can lead to the degradation of METTL14 and METTL3, thereby reducing the level of m6A and affecting the regulation of gene expression (38), since WTAP was first found to be highly expressed in glioblastoma and an essential indicator of poor prognosis (56), more and more studies have found that it is abnormally said in a variety of malignant tumors and affects the survival and prognosis of patients. For example, the overexpression of WTAP in cholangiocarcinoma, especially in lymph nodes or vascular metastatic cholangiocarcinoma cells, can significantly increase tumor cells’ migration and invasion ability. However, its involvement as an m6A modifying enzyme in carcinogenesis has not been elucidated (57). Li et al. found that WTAP is highly expressed in gastric cancer, and its carcinogenic effect may be related to the high methylation of RNA of T-cell immune response-related genes (58). WTAP can down-regulate the expression of CAV-1 by regulating the m6A modification of CAV-1 mRNA, thereby activating the NF-κB signaling pathway to promote the progression of endometrial cancer (59). In addition to solid tumors, WTAP promotes diffuse large B-cell lymphoma (DLBCL). Kuai et al. found that WTAP was highly expressed in DLBCL and demonstrated that WTAP formed stable complexes with heat shock protein 90 (Hsp90) and B-cell lymphoma six protein (Bcl-6). Thus, the proliferation and anti-apoptosis ability of DLBCL cells were promoted (60). WTAP can also regulate the expression of HMBOX1 in an m6A-dependent manner and promote the progression and metastasis of osteosarcoma through PI3K/AKT signaling pathway (61). WTAP depends not only on m6A to participate in the occurrence and development of tumors but is also closely related to other diseases. For example, WTAP can increase endoplasmic reticulum stress and apoptosis by regulating m6A modification of ATF4 mRNA, promoting myocardial ischemia and reperfusion injury (62), WTAP-mediated m6A modification of lncRNA NORAD promotes nucleus pulposus cell senescence and intervertebral disc degeneration (63). As an essential component of the m6A methyltransferase complex, WTAP not only participates in the occurrence, development, and metastasis of various malignant tumors by regulating m6A methylation modification but also plays an essential role in the development of non-neoplasia diseases such as renal injury (64), myocardial injury and intervertebral disc degeneration.
Secondly, the abnormal modification of m6A mediated by WTAP can affect cancer occurrence, development, and metastasis by reshaping TME. WTAP is significantly associated with the event and product of cancer and the poor prognosis of patients, but the specific mechanism of action is still unclear. As a virtual environment for tumor survival, TME plays an essential role in the tumor progression process and affects immunotherapy (65). Increasing evidence indicates that m6A modification in tumor cells is closely related to immune microenvironment. On the one hand, m6A can not only regulate the differentiation and function of immune cells in the immune microenvironment, such as macrophage polarization and T-cell activation, but also regulate the expression of immune cytokines, such as the production of interferon and tumor necrosis factor affecting the interaction between immune cells and the immune regulatory process (19). On the other hand, m6A can also directly regulate the transcription and translation of immune-related genes and affect the immune response. In head and neck squamous cell carcinoma (HNSCC), PD-L1 expression and immune cell infiltration in TME significantly correlate with m6A regulators (66). Bioinformatics analysis of nasopharyngeal carcinoma (NPC) showed that m6A methylation was associated with immune infiltration and escape in TME (67). Tang et al. investigated the relationship between the mutation status of m6A-related genes and the tumor immune microenvironment of pancreatic cancer (PAAD), showing that m6A modification may be involved in immune cell infiltration, and the composition of infiltrating immune cells may affect m6A modification (68). It was found that m6A modification is also involved in forming TME and immune-related pathways in lung adenocarcinoma (LUAD) (69). WTAP, as a methyltransferase, has been confirmed to regulate m6A methylation, and m6A modification is closely related to the infiltration of immune cells in TME; m6A modification can affect the abundance of immune cell infiltration in TME through a variety of signaling pathways, thus affecting the biological behavior of tumors (70). This paper also preliminarily verified that as an essential regulator of m6A methylation, WTAP is highly expressed in many tumors and closely related to TME remodeling. Therefore, we speculate that WTAP may reshape the TME through abnormal modification of m6A, thus affecting cancer’s occurrence, development, and survival prognosis, but its specific mechanism needs further experimental studies.
The third is the application prospect of m6A and WTAP as potential therapeutic targets in cancer. As the most extensive modification of eukaryotic mRNA, m6A links epitranscriptomics to the occurrence and development of tumors, affecting self-renewal and differentiation of tumor stem cells, proliferation and apoptosis (37), drug resistance (71), immunosuppression (72) and other processes. m6A methylation and demethylation modification is a reversible process. The level of m6A in cells can be changed by promoting or inhibiting some critical enzymes in the process of m6A modification to achieve the purpose of intervening in tumor progression. Huang et al. found that meclofenamic acid (MA) could inhibit the m6A demethylase activity of FTO, performing a selective chemical intervention in RNA demethylation (73). Subsequently, the ethyl ester derivative MA2 of MA has been approved for clinical treatment in the United States and has shown promising efficacy in glioma models (74). In 2019, Huang et al. reported two potent small-molecule FTO inhibitors (FB23 and FB23-2), which directly bind to FTO and selectively inhibit the m6A demethylase activity of FTO, promote apoptosis and inhibit the proliferation of AML cells (75). Visvanathan et al. found that silencing METTL3 could inhibit glioblastoma growth by enhancing tumor sensitivity to radiotherapy (76). In addition, baicalin hydrate can promote splicing of histone methylase SUV39H1 by enhancing m6A methylation, induce cell cycle arrest and apoptosis, and play a role in inhibiting the proliferation of nasopharyngeal carcinoma cells (77). Since understanding m6A modification and the regulatory mechanism of key enzymes needs to be further studied, the current achievements mainly focus on the selective inhibitors of FTO, and the therapeutic effect requires more clinical verification. Many studies have shown that WTAP plays an essential role in tumorigenesis and development and may also promote tumor cell apoptosis, immunosuppression, and drug resistance as potential therapeutic targets. For pediatric tumors, such as leukemia and osteosarcoma, WTAP-related studies have contributed to a better understanding of their molecular mechanisms and developmental processes. By studying the expression level and function of WTAP in pediatric tumors and its relationship with prognosis, we can explore its potential as a prognostic assessment for pediatric tumors. These findings may help clinicians to develop better individualized treatment plans to improve the therapeutic efficacy and survival rate of pediatric tumors. For the aspect of adult tumors, the related studies of WTAP are also of great significance. Understanding the expression pattern, function, and relationship with prognosis of WTAP in different types of adult tumors can help us better understand the different subtypes and molecular features of adult tumors. This has important clinical implications for the development of precise treatment strategies and the prediction of patient response to immunotherapy. Currently, targeted drug therapy and immunotherapy for WTAP are still developing. However, with the deepening of RNA methylation research, the application of WTAP-related targeted drugs, as well as the development of small-molecule inhibitors targeting m6A regulatory proteins and their novel drugs in combination with immune checkpoint blockers are bound to play an important value in the future.
TME and immune infiltration analysis
TME is closely related to the occurrence and development of tumors. Understanding the microenvironment of tumors, including the infiltration of immune cells, will help to explore the mechanism of tumor development further and provide ideas for novel cancer treatment. WTAP is associated with the occurrence and development of various tumors and may affect the survival and prognosis of patients. To further analyze the expression difference of WTAP in tumor tissues and explore its predictive value for tumor prognosis. We performed gene expression differential analysis and prognosis analysis. The results showed that WTAP was significantly differentially expressed between most tumors and adjacent normal tissues, and the expression of WTAP had an essential impact on the survival rate of cancer patients. High expression of WTAP was negatively correlated with the overall survival rate in various tumors. To investigate whether WTAP is involved in regulating tumor-related immune responses, we performed immune cell analysis, immune infiltration analysis, and immune checkpoint analysis. The results showed that the expression of WTAP was significantly correlated with immune infiltration in most tumors, and WTAP was correlated considerably with TME in many cancers and was most strongly associated with stromal cell scores in GBMLGG, KIPAN, and PAAD, and with immune cell scores in GBMLGG, KIPAN and DLBC. The strongest correlation was found with the ESTIMATE scores of GBMLGG, KIPAN, and DLBC. These results suggest that WTAP is highly involved in TME remodeling in these tumors. At the same time, in the ESTIMATE score, we observed that WTAP expression was significantly correlated with immune infiltration in 25 cancer species, of which nineteen were significantly positively correlated, and six were significantly negatively correlated. The relationship between WTAP and immune checkpoint gene expression showed that the expression level of WTAP in each tumor was associated with more than 40 checkpoints, most of which were positively correlated. It has also been shown that inhibition of m6A modification can increase tumor sensitivity to immunotherapy by altering TME and CD8+ T cell recruitment, revealing the function of RNA methylation in adaptive immunity (78).
All these immune correlation analyses suggest that WTAP may be essential as a potential biomarker in tumor immunity. Tumor mutation burden (TMB) and microsatellite instability (MSI) are new tumors immunotherapy prediction markers. Studies have shown that they can guide the clinical treatment of patients with non-small cell lung cancer (79) and colon adenocarcinoma (80). This study found that the expression of WTAP was significantly correlated with TMB of 11 cancers, and the strongest correlation was found with GBMLGG (R=0.41). WTAP was connected considerably with MSI of 12 cancers, and DLBC (R=0.54) had the strongest correlation. Therefore, WTAP is considered an important indicator to evaluate the efficacy of immunotherapy, and it can further regulate tumor immunity by affecting TMB and MSI and becoming a new target in immunotherapy. In conclusion, this study found that WTAP is associated with various cancers’ TMEs and immune infiltration, especially GBMLGG, KIPAN, and DLBC. It is expected to become a breakthrough for new cancer immunotherapy and targeted therapy.
Limitations of this study
This is the first bibliometric analysis of WTAP-related research areas, and there are inherent limitations in using CiteSpace and VOSviewer for visual analysis. Firstly, to meet the reference format requirements of the two software, data retrieval was conducted only from the Web of Science core collection database, leaving out the literature not included in the database (26). Secondly, some keywords extracted based on software have no prominent discussion and research significance, so it is necessary to combine the researcher’s comprehensive understanding of keywords in the research field to select, so there may be deviation (27). As for the first limitation, WoSCC, the most commonly used scientometric analysis database, is one of the most authoritative scientific reference retrieval tools, representing most of the information (26). However, only a few reference data are not included, which will not significantly impact bibliometric analysis. As for the second limitation, for the possible deviation caused by the choice of keywords, researchers must read a large number of relevant references, and cultivate rigorous scientific research logical thinking ability, to minimize the subjective deviation.
Conclusions
As an essential component of the m6A methyltransferase complex, WTAP participates in the occurrence and development of many diseases, especially tumors, by regulating m6A methylation modification. Currently, the research on WTAP is in the stage of rapid growth, and the cooperation between the countries, institutions, and authors is close. Although the research on WTAP has been preliminarily recognized, there are still many problems to be solved, so it is imperative to carry out further research in this field. The bibliometric analysis results show that current WTAP research’s main aspects include m6A modification, tumor, cancer treatment, and regulatory mechanisms.
The research frontiers and hotspots are m6A modification, methyltransferase, demethylation, TME, and immunotherapy. The results of bioinformatics analysis showed that the expression of WTAP was up-regulated in various tumors and affected patients’ survival prognoses. WTAP was associated with remodeling tumor immune microenvironment in multiple cancers, which is expected to become a potential molecular target for cancer treatment and drug development. Based on these results, the emerging topics will be closely related to the basic mechanism research and clinical application research of m6A and WTAP-related diseases and explore the in-depth mechanism research of WTAP through abnormal m6A modification and remodeling of TME to promote the occurrence, development, and metastasis of cancer. This paper can provide relevant researchers with research hotspots and frontier trends in this field and reference ideas for finding new topics and directions.
Acknowledgments
The authors thank the Third Xiangya Hospital of Central South University for their support of this work and the reviewers for allowing us to make improvements to the manuscript.
Funding: This study was supported by
Footnote
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1243/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1243/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum Mol Genet 2000;9:2231-9. [Crossref] [PubMed]
- Li YL, Yu J, Song SH. Recent progresses in RNA N6-methyladenosine research. Yi Chuan 2013;35:1340-51. [Crossref] [PubMed]
- Zhang Y, Geng X, Li Q, et al. m6A modification in RNA: biogenesis, functions and roles in gliomas. J Exp Clin Cancer Res 2020;39:192. [Crossref] [PubMed]
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, et al. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem 1994;269:17697-704. [Crossref] [PubMed]
- Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997;3:1233-47. [PubMed]
- Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014;24:177-89. [Crossref] [PubMed]
- Ito-Kureha T, Leoni C, Borland K, et al. The function of Wtap in N(6)-adenosine methylation of mRNAs controls T cell receptor signaling and survival of T cells. Nat Immunol 2022;23:1208-21. [Crossref] [PubMed]
- Horiuchi K, Kawamura T, Hamakubo T. Wilms’ tumor 1-associating protein complex regulates alternative splicing and polyadenylation at potential G-quadruplex-forming splice site sequences. J Biol Chem 2021;297:101248. [Crossref] [PubMed]
- Cui Q, Shi H, Ye P, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 2017;18:2622-34. [Crossref] [PubMed]
- Weng H, Huang H, Wu H, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 2018;22:191-205.e9. [Crossref] [PubMed]
- Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. [Crossref] [PubMed]
- Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017;65:529-43. [Crossref] [PubMed]
- Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Parkin J, Cohen B. An overview of the immune system. Lancet 2001;357:1777-89. [Crossref] [PubMed]
- Roma-Rodrigues C, Mendes R, Baptista PV, et al. Targeting Tumor Microenvironment for Cancer Therapy. Int J Mol Sci 2019;20:840. [Crossref] [PubMed]
- Affo S, Yu LX, Schwabe RF. The Role of Cancer-Associated Fibroblasts and Fibrosis in Liver Cancer. Annu Rev Pathol 2017;12:153-86. [Crossref] [PubMed]
- Huai Q, Guo W, Han L, et al. Identification of prognostic genes and tumor-infiltrating immune cells in the tumor microenvironment of esophageal squamous cell carcinoma and esophageal adenocarcinoma. Transl Cancer Res 2021;10:1787-803. [Crossref] [PubMed]
- Li X, Ma S, Deng Y, et al. Targeting the RNA m6A modification for cancer immunotherapy. Mol Cancer 2022;21:76. [Crossref] [PubMed]
- Gu Y, Wu X, Zhang J, et al. The evolving landscape of N6-methyladenosine modification in the tumor microenvironment. Mol Ther 2021;29:1703-15. [Crossref] [PubMed]
- Smith DR. Bibliometrics, dermatology and contact dermatitis. Contact Dermatitis 2008;59:133-6. [Crossref] [PubMed]
- Chen C. Searching for intellectual turning points: progressive knowledge domain visualization. Proc Natl Acad Sci U S A 2004;101:5303-10. [Crossref] [PubMed]
- van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010;84:523-38. [Crossref] [PubMed]
- Mulet-Forteza C, Genovart-Balaguer J, Mauleon-Mendez E, et al. A bibliometric research in the tourism, leisure and hospitality fields. Journal of Business Research 2019;101:819-27. [Crossref]
- Qin Y, Zhang Q, Liu Y. Analysis of knowledge bases and research focuses of cerebral ischemia-reperfusion from the perspective of mapping knowledge domain. Brain Res Bull 2020;156:15-24. [Crossref] [PubMed]
- Zhang J, Song L, Xu L, et al. Knowledge Domain and Emerging Trends in Ferroptosis Research: A Bibliometric and Knowledge-Map Analysis. Front Oncol 2021;11:686726. [Crossref] [PubMed]
- Ai Y, Xing Y, Yan L, et al. Atrial Fibrillation and Depression: A Bibliometric Analysis From 2001 to 2021. Front Cardiovasc Med 2022;9:775329. [Crossref] [PubMed]
- Ge Y, Chao T, Sun J, et al. Frontiers and Hotspots Evolution in Psycho-cardiology: A Bibliometric Analysis From 2004 to 2022. Curr Probl Cardiol 2022;47:101361. [Crossref] [PubMed]
- Wu H, Cheng K, Tong L, et al. Knowledge structure and emerging trends on osteonecrosis of the femoral head: a bibliometric and visualized study. J Orthop Surg Res 2022;17:194. [Crossref] [PubMed]
- Miao L, Zhang J, Zhang Z, et al. A Bibliometric and Knowledge-Map Analysis of CAR-T Cells From 2009 to 2021. Front Immunol 2022;13:840956. [Crossref] [PubMed]
- Mulet-Forteza C, Genovart-Balaguer J, Merigó J M, et al. Bibliometric structure of IJCHM in its 30 years. International Journal of Contemporary Hospitality Management 2019;31:4574-604. [Crossref]
- Chen C, Hu Z, Liu S, et al. Emerging trends in regenerative medicine: a scientometric analysis in CiteSpace. Expert Opin Biol Ther 2012;12:593-608. [Crossref] [PubMed]
- Chen C, Leydesdorff L. Patterns of connections and movements in dual-map overlays: A new method of publication portfolio analysis. Journal of the Association for Information Science and Technology 2014;65:334-51. [Crossref]
- Chen C. Science Mapping: A Systematic Review of the Literature Journal of Data and Information Science 2017;2:1-40. [J]. [Crossref]
- Gao Y, Shi S, Ma W, et al. Bibliometric analysis of global research on PD-1 and PD-L1 in the field of cancer. Int Immunopharmacol 2019;72:374-84. [Crossref] [PubMed]
- Ma D, Yang B, Guan B, et al. A Bibliometric Analysis of Pyroptosis From 2001 to 2021. Front Immunol 2021;12:731933. [Crossref] [PubMed]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. [Crossref] [PubMed]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93-5. [Crossref] [PubMed]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18-29. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep 2014;8:284-96. [Crossref] [PubMed]
- Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016;537:369-73. [Crossref] [PubMed]
- Horiuchi K, Kawamura T, Iwanari H, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 2013;288:33292-302. [Crossref] [PubMed]
- Bansal H, Yihua Q, Iyer SP, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 2014;28:1171-4. [Crossref] [PubMed]
- Xiao F, Li C, Sun J, et al. Knowledge Domain and Emerging Trends in Organic Photovoltaic Technology: A Scientometric Review Based on CiteSpace Analysis. Front Chem 2017;5:67. [Crossref] [PubMed]
- Liu G, Jiang R, Jin Y. Sciatic nerve injury repair: a visualized analysis of research fronts and development trends. Neural Regen Res 2014;9:1716-22. [Crossref] [PubMed]
- Chen CM, Dubin R, Kim MC. Orphan drugs and rare diseases: a scientometric review (2000-2014). Exp Opin Orphan Drugs 2014;2:709-24. [Crossref]
- Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res 2018;28:507-17. [Crossref] [PubMed]
- Yang S, Wei J, Cui YH, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun 2019;10:2782. [Crossref] [PubMed]
- Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer 2019;18:103. [Crossref] [PubMed]
- Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer 2019;18:112. [Crossref] [PubMed]
- Zhang J, Tsoi H, Li X, et al. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut 2016;65:1482-93. [Crossref] [PubMed]
- Wen J, Lv R, Ma H, et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell 2018;69:1028-1038.e6. [Crossref] [PubMed]
- Yu HL, Ma XD, Tong JF, et al. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. Onco Targets Ther 2019;12:6191-201. [Crossref] [PubMed]
- Van Nieuwenhuysen E, Busschaert P, Neven P, et al. The genetic landscape of 87 ovarian germ cell tumors. Gynecol Oncol 2018;151:61-8. [Crossref] [PubMed]
- Jin DI, Lee SW, Han ME, et al. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci 2012;103:2102-9. [Crossref] [PubMed]
- Jo HJ, Shim HE, Han ME, et al. WTAP regulates migration and invasion of cholangiocarcinoma cells. J Gastroenterol 2013;48:1271-82. [Crossref] [PubMed]
- Li H, Su Q, Li B, et al. High expression of WTAP leads to poor prognosis of gastric cancer by influencing tumour-associated T lymphocyte infiltration. J Cell Mol Med 2020;24:4452-65. [Crossref] [PubMed]
- Li Q, Wang C, Dong W, et al. WTAP facilitates progression of endometrial cancer via CAV-1/NF-κB axis. Cell Biol Int 2021;45:1269-77. [Crossref] [PubMed]
- Kuai Y, Gong X, Ding L, et al. Wilms’ tumor 1-associating protein plays an aggressive role in diffuse large B-cell lymphoma and forms a complex with BCL6 via Hsp90. Cell Commun Signal 2018;16:50. [Crossref] [PubMed]
- Chen S, Li Y, Zhi S, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m6A-dependent manner. Cell Death Dis 2020;11:659. [Crossref] [PubMed]
- Wang J, Zhang J, Ma Y, et al. WTAP promotes myocardial ischemia/reperfusion injury by increasing endoplasmic reticulum stress via regulating m6A modification of ATF4 mRNA. Aging (Albany NY) 2021;13:11135-49. [Crossref] [PubMed]
- Li G, Ma L, He S, et al. WTAP-mediated m6A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat Commun. 2022;13(1):1469.Correction appears in Nat Commun 2022;13:3572.
- Lan J, Xu B, Shi X, et al. WTAP-mediated N6-methyladenosine modification of NLRP3 mRNA in kidney injury of diabetic nephropathy. Cell Mol Biol Lett 2022;27:51. [Crossref] [PubMed]
- Tang H, Qiao J, Fu YX. Immunotherapy and tumor microenvironment. Cancer Lett 2016;370:85-90. [Crossref] [PubMed]
- Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive Analysis of the PD-L1 and Immune Infiltrates of m6A RNA Methylation Regulators in Head and Neck Squamous Cell Carcinoma. Mol Ther Nucleic Acids 2020;21:299-314. [Crossref] [PubMed]
- Lu S, Yu Z, Xiao Z, et al. Gene Signatures and Prognostic Values of m6A Genes in Nasopharyngeal Carcinoma. Front Oncol 2020;10:875. [Crossref] [PubMed]
- Tang R, Zhang Y, Liang C, et al. The role of m6A-related genes in the prognosis and immune microenvironment of pancreatic adenocarcinoma. PeerJ 2020;8:e9602. [Crossref] [PubMed]
- Li Y, Gu J, Xu F, et al. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform 2021;22:bbaa225. [Crossref] [PubMed]
- Li M, Zha X, Wang S. The role of N6-methyladenosine mRNA in the tumor microenvironment. Biochim Biophys Acta Rev Cancer 2021;1875:188522. [Crossref] [PubMed]
- Wei W, Sun J, Zhang H, et al. Circ0008399 Interaction with WTAP Promotes Assembly and Activity of the m6A Methyltransferase Complex and Promotes Cisplatin Resistance in Bladder Cancer. Cancer Res 2021;81:6142-56. [Crossref] [PubMed]
- Xiong J, He J, Zhu J, et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell 2022;82:1660-77.e10. [Crossref] [PubMed]
- Huang Y, Yan J, Li Q, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res 2015;43:373-84. [Crossref] [PubMed]
- Zhang S, Zhao BS, Zhou A, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017;31:591-606.e6. [Crossref] [PubMed]
- Huang Y, Su R, Sheng Y, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019;35:677-691.e10. [Crossref] [PubMed]
- Visvanathan A, Patil V, Arora A, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018;37:522-33. [Crossref] [PubMed]
- Lai W, Jia J, Yan B, et al. Baicalin hydrate inhibits cancer progression in nasopharyngeal carcinoma by affecting genome instability and splicing. Oncotarget 2017;9:901-14. [Crossref] [PubMed]
- Wang L, Hui H, Agrawal K, et al. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J 2020;39:e104514. [Crossref] [PubMed]
- Shim JH, Kim HS, Cha H, et al. HLA-corrected tumor mutation burden and homologous recombination deficiency for the prediction of response to PD-(L)1 blockade in advanced non-small-cell lung cancer patients. Ann Oncol 2020;31:902-11. [Crossref] [PubMed]
- Allan RE, Luis RP, Juan P. Microsatellite instability in Costa Rican patients with colorectal adenocarcinoma and its association with overall survival and response to fluoropyrimidine-based chemotherapy. Cancer Epidemiol 2020;65:101680. [Crossref] [PubMed]


