Non-targeted metabolomics analysis of indoleamine 2,3-dioxygenase inhibitor treatment in a mouse model of early-stage lung adenocarcinoma
Highlight box
Key findings
• Early-stage lung adenocarcinoma-specific biomarkers were identified using non-targeted metabolomics analysis in an established mouse model.
What is known and what is new?
• Early-stage lung adenocarcinoma often lacks obvious symptoms, making accurate diagnosis challenging if based on clinical presentations alone.
• Metabolic alterations associated with early-stage lung adenocarcinoma in mice were revealed.
What is the implication, and what should change now?
• This study provides clues for further investigation into the early diagnosis and treatment of lung adenocarcinoma. By gaining a deeper understanding of the biological significance of the differential metabolites, we may discover new biomarkers or therapeutic targets, providing better strategies for personalized treatment and management of lung adenocarcinoma.
Introduction
Lung cancer is one of the most common cancers with high incidence and mortality rates (1). The incidence rate of lung cancer ranks second among both male and female cancer cases, reaching 14% and 13% respectively, while the proportion of lung cancer-related deaths ranks first among both male and female cancer-related deaths, reaching 26% and 25% respectively (2). In China, lung cancer has the highest incidence rate among all malignant tumors (3). According to pathological classification, lung cancer is mainly divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). Among them, lung adenocarcinoma, which belongs to NSCLC, is the most common type of lung cancer in China, accounting for more than two-thirds of lung cancer patients, with a higher incidence in females (3). However, clinical studies have shown that the five-year survival rate after surgery for early-stage lung adenocarcinoma can reach 90%, while for patients in the middle and late stages, the survival rate decreases significantly due to metastasis of lung adenocarcinoma cells, making surgical treatment no longer feasible and necessitating a combination of radiotherapy and chemotherapy, resulting in a poor prognosis (4,5). This is likely due to the fact that lung adenocarcinoma can infiltrate and grow into the bronchial cavity, adjacent lung tissue, and other organs, as well as spreading to other sites through lymphatic and hematogenous routes. Therefore, most patients are difficult to be diagnosed at an early stage and are often reached when detected, leading to a poor prognosis (6).
Immunotherapy is a primary treatment modality for patients with lung adenocarcinoma, especially for those with sensitive genes without mutations, resistance to oral targeted drugs, and resistance to conventional chemotherapy. Large molecule biological agents such as antibodies have been applied in clinical research and in clinical settings (7), while small molecule inhibitors have gradually become an important research focus in tumor immunotherapy due to their relatively simple administration and lower development costs. Indoleamine 2,3-dioxygenase (IDO) is an extrahepatic rate-limiting enzyme involved in tryptophan metabolism in the human body, and it can regulate the body’s anti-tumor immune response by mediating tryptophan depletion and its metabolic products (8,9). IDO is overexpressed in a wide range of cancer cells, leading to immune escape of cancer cells, such as the case in lung cancer, pancreatic cancer, breast cancer, colorectal cancer, and others (10). Previous study has shown that IDO is closely related to the pathogenesis of various tumors and may become a key enzyme in tumor treatment (11). Epacadostat, an effective and selective IDO inhibitor, has been developed as an antitumor drug and has entered phase II clinical trials (12,13). However, there is limited research on the treatment of lung adenocarcinoma with Epacadostat.
Metabolomics is a high-throughput, high-resolution mass spectrometry analysis technique that primarily focuses on analyzing the final metabolites in a biological system to identify the metabolic pathways and changes in differential metabolites in response to external stimuli (14). In recent years, metabolomics has been applied in the early diagnosis of various cancers and has shown good sensitivity and specificity (15,16). Furthermore, metabolomics has significant advantages in the screening and identification of tumor biomarkers (17-19). Wu et al. (20) analyzed gastric mucosal tissue samples from 18 gastric cancer patients using gas chromatography-mass spectrometry and established a gastric cancer diagnostic model using principal component analysis (PCA), which was evaluated by receiver operating characteristic curves. The results revealed 18 different metabolites present in malignant and non-malignant tissues. Fan et al. (21) used ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS) to identify small molecular metabolites that could serve as potential indicators for ovarian cancer screening. In addition, metabolomics has also been applied in lung cancer. Hu et al. (22) analyzed the plasma of healthy controls and NSCLC patients using nuclear magnetic resonance (NMR)-based metabolomics analysis, and found that the concentrations of lactate, alanine, and other metabolites were significantly elevated in the plasma of NSCLC patients compared to the normal group, while glucose and taurine concentrations were significantly decreased. Carrola et al. (23) also used NMR-based metabolomics analysis to study the changes in metabolic profiles in urine samples from lung cancer patients, and found decreased concentrations of hippurate and scyllo-inositol, as well as increased concentrations of β-hydroxyisovalerate, α-hydroxyisovalerate, N-acetylglutamine, and creatinine compared to the normal group. In addition to the studies mentioned above, many other studies have also investigated metabolite changes in lung cancer patients in urine and plasma samples. For example, Kawamoto et al. (24) discovered specific metabolite markers associated with lung cancer progression, while Han et al. (25) used a metabolomics approach to reveal mutated genes that affect metabolic differences between lung cancer and healthy individuals.
In this study, an early-stage lung adenocarcinoma mouse model was established by subcutaneous injection of nitrosourea. The UPLC-QTOF-MS metabolomics analysis method was used to analyze the profile changes of metabolites in mouse plasma and urine samples. At the same time, we performed epacadostat intervention therapy on lung adenocarcinoma mice and analyzed the changes in metabolites in plasma and urine samples after intervention, as well as the changes in metabolic pathways related to metabolites, in order to explore the intervention mechanism of epacadostat in the treatment of early-stage lung adenocarcinoma in mice. We present this article in accordance with the ARRIVE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1236/rc).
Methods
Early-stage lung adenocarcinoma modeling
Twenty specific-pathogen-free-grade healthy 4-week-old female Kunming mice were purchased from SPF Biotechnology Company (Beijing, China) and were used to establish the early-stage lung adenocarcinoma mouse model using the subcutaneous injection of nitrosourea solution method. The mice were kept in a clean environment and were subcutaneously injected with 0.2 mL of nitrosourea solution at a concentration of 2.0 mg/mL for four consecutive weeks. The model was established after 100 days. The animal experiments were conducted with the approval from the Ethics Committee of the First Affiliated Hospital, University of South China (No. 2019LL1021001), in compliance with national or institutional guidelines for the care and use of animals.
Experimental grouping
All mice were divided into a normal control group and an experimental group. The normal control group consisted of 5 mice, which were raised conventionally for 100 days without intervention. The experimental group mice were randomly divided into three groups, with 5 mice in each group. The early-stage lung adenocarcinoma group consisted of 5 mice without any intervention at any stages of the course. The epacadostat intervention group consisted of 10 mice, and after the establishment of the early-stage lung adenocarcinoma model, the mice were orally administered epacadostat suspension twice a day for 28 consecutive days. The epacadostat intervention group was further divided into Epacadostatlow group and Epacadostathigh group, with concentrations of 20 and 500 mg/kg, respectively [epacadostat was prepared in 1 mL of 0.5% carboxymethylcellulose sodium (CMC-Na)].
Collection and processing of tissue specimens
At two time points, 7 and 28 days, blood samples and urine samples were collected from the 20 mice. Samples collected at 7 days were labeled as T7 for the experimental group and C7 for the control group. The Epacadostatlow group was labeled as TL7, and the Epacadostathigh group was labeled as TH7. Samples collected at 28 days were labeled as T28 for the experimental group and C28 for the control group. The Epacadostatlow group was labeled as TL28, and the Epacadostathigh group was labeled as TH28. Plasma and urine samples were stored in a −80 ℃ freezer and transported on dry ice. Plasma samples were thawed at room temperature, and 200 µL of plasma was mixed with 800 µL of four-fold acetonitrile, vortexed for 30 s, and centrifuged at 12,000 rpm for 15 min at 4 ℃. The supernatant was collected, and the pellet was discarded. The supernatant was evaporated in a centrifugal concentrator for 2 h, and the residue was dissolved in 200 µL of 10% acetonitrile solution, vortexed for 1 min, and centrifuged at 12,000 rpm for 15 min at 4 ℃. The supernatant was used for UPLC-QTOF-MS analysis. Urine samples were thawed at room temperature, and 200 µL of urine was placed in a centrifuge tube and centrifuged at 12,000 rpm for 15 min at 4 ℃. The supernatant was collected, and the subsequent procedures were the same as the plasma sample processing.
UPLC-QTOF-MS analysis
The UPLC-QTOF-MS analysis was performed at Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China). The raw data collected was converted into mzXML format using ProteoWizard, and then peak alignment, retention time correction, and peak area extraction were performed using XCMS software (VenusBio Technology Co., Ltd., Guangzhou, China). The data extracted by XCMS was firstly subjected to metabolite structure identification and data preprocessing, followed by an assessment of the quality of experimental data, and finally, data analysis was conducted. The ion fragmentation spectra were obtained through UPLC-QTOF-MS secondary mass spectrometry analysis and compared with the obtained spectra in the experiment for confirmation.
Bioinformatics analysis
The expression levels of all samples and differentially expressed metabolites were calculated using a distance matrix, and hierarchical clustering was performed for cluster analysis. Correlation analysis was used to measure the metabolic associations between significantly different metabolites [variable importance in the projection (VIP) >1, P value <0.05] and understand the interregulation of metabolites during the process of biological changes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to evaluate the metabolic pathway enrichment of differentially expressed metabolites (https://www.genome.jp/kegg/pathway.html). Metabolic pathways with a P value <0.05 were considered significantly enriched.
Statistical analysis
For data pre-processing, raw metabolomics data were subjected to normalization to account for differences in sample concentrations. Peaks were aligned across samples, and the intensities were scaled using a standard internal control. Handling of missing values was carried out using imputation techniques. For metabolites with less than 20% missing values, the missing data were imputed using the k-nearest neighbors algorithm. For metabolites with more than 20% missing values, they were removed from the dataset to ensure robust statistical analysis. After normalized to total peak intensity, the processed data were analyzed by R package (ropls), where it was subjected to multivariate data analysis, including Pareto-scaled PCA and orthogonal partial least squares discriminant analysis (OPLS-DA). The 7-fold cross-validation and response permutation testing were used to evaluate the robustness of the model. The VIP value of each variable in the OPLS-DA model was calculated to indicate its contribution to the classification. Metabolites with the VIP value >1 was further applied to Student’s t-test at univariate level to measure the significance of each metabolite, the P values less than 0.05 were considered as statistically significant.
Results
Metabolite screening
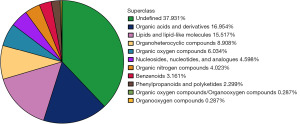
The quantitative results of metabolites were displayed in positive and negative ion modes. Quality control (QC) samples were analyzed using total ion chromatograms (TIC) and PCA to assess their stability and accuracy. The results shown in Figure S1 demonstrate that the instrument was stable and the data were reliable. A total of 348 metabolites were identified, with 237 metabolites identified in the positive ion mode and 111 metabolites identified in the negative ion mode after merging the data. Excluding undefined metabolites, organic acids and derivatives (16.954%) and lipids and lipid-like molecules (15.517%) were the two most abundant metabolite classes in early-stage lung adenocarcinoma mice (Figure 1).
Differential metabolite analysis in early-stage lung adenocarcinoma mice at day 7
The results of univariate statistical analysis were visually presented in volcano plots, indicating differentially expressed metabolites in the positive and negative ion modes between groups C7 and T7 (Figure S2A,S2B), T7 and TL7 (Figure S2C,S2D), and TL7 and TH7 (Figure S2E,S2F). OPLS-DA analysis was performed to maximize the differentiation between groups, including intergroup and intragroup variations. The results showed significant differences between groups C7 and T7 (Figure 2A,2B), T7 and TL7 (Figure 2C,2D), and TL7 and TH7 (Figure 2E,2F). In the positive ion mode, T7 exhibited greater intragroup differences compared to C7. In the negative ion mode, T7 exhibited larger intragroup differences compared to TL7. In the positive ion mode, TL7 exhibited larger intragroup differences compared to TH7. OPLS-DA VIP >1 and P value <0.05 were used as thresholds to select significantly different metabolites. The comparison between T7 and C7 in the negative ion mode revealed high expression differences in ethyl glucuronide, belonging to organic oxygen compounds, and low expression differences in glyceric acid (Figure 3A). In the positive ion mode, anthranilic acid (vitamin L1), belonging to benzenoids, showed high expression differences (Figure 3B). The comparison between T7 and TL7 in the negative ion mode showed low expression differences in dodecanoic acid and 2-ethyl-2-hydroxybutyric acid, belonging to lipids and lipid-like molecules (Figure 3C). In the positive ion mode, hydrocortisone (cortisol), belonging to lipids and lipid-like molecules, showed high expression differences (Figure 3D). The comparison between TL7 and TH7 in the negative ion mode revealed low expression differences in hippuric acid, belonging to benzenoids (Figure 3E). In the positive ion mode, anthranilic acid (vitamin L1), belonging to benzenoids, showed high expression differences (Figure 3F).


Differential metabolite analysis in early-stage lung adenocarcinoma mice at day 28
Volcano plots were used to intuitively represent the results of univariate statistical analysis, showing differentially expressed metabolites between the C28 and T28 groups (Figure S3A,S3B), T28 and TL28 groups (Figure S3C,S3D), and TL28 and TH28 groups (Figure S3E,S3F) in both positive and negative ion modes. OPLS-DA was employed to maximize the differences in metabolites between groups, encompassing both intergroup and intragroup variations. The results indicated significant differences between groups: C28 and T28 (Figure 4A,4B), T28 and TL28 (Figure 4C,4D), and TL28 and TH28 (Figure 4E,4F). In both positive and negative ion modes, the intragroup differences in the T28 group were more pronounced compared to the C28 group. Compared to the T28 group, the TL28 group exhibited larger intragroup discrepancies in both ion modes. In the positive ion mode, the TL28 group showed greater intragroup variation compared to the TH28 group. Metabolites with OPLS-DA VIP >1 and P value <0.05 were selected as significant. The comparison between the T28 and C28 groups revealed that benzoic acid, belonging to benzenoids, was downregulated in the negative ion mode (Figure 5A). In the positive ion mode, 1-methylhistidine, classified under organic acids and derivatives, was downregulated (Figure 5B). Comparing the T28 and TL28 groups, dodecanoic acid, a lipid-like molecule, showed lower expression in the negative ion mode (Figure 5C). In the positive ion mode, N-acetyl-L-alanine and taurine, both organic acids and derivatives, were upregulated, whereas N-(omega)-hydroxyarginine was downregulated (Figure 5D). The comparison between the TL28 and TH28 groups revealed that 1,4-dihydroxybenzene and hippuric acid, both benzenoids, were downregulated in the negative ion mode (Figure 5E). In the positive ion mode, anthranilic acid (vitamin L1), also a benzenoid, was upregulated (Figure 5F).


Differential metabolite clustering and correlation analysis
Bioinformatics analysis was performed on the selected significant differential metabolites. Hierarchical clustering analysis was conducted to visually and comprehensively display the relationship between samples and the expression pattern differences of metabolites in different samples. At day 7, in the positive ion mode, 9 metabolites including urea, 1-myristoyl-sn-glycero-3-phosphocholine, and 1-methylhistidine were found to have intergroup differences between groups C7 and T7 (Figure S4A). The correlation analysis results showed positive correlations between anthranilic acid (vitamin L1) and Pro-Met, 1-methylhistidine, and creatinine, and a negative correlation with procaterol (Figure 6A). In the negative ion mode, 7 differential metabolites between groups TH7 and TL7 were displayed (Figure S4B). The correlation analysis results showed a positive correlation between hippuric acid and 12(R)-HETE (Figure 6B). In the positive ion mode, 10 differential metabolites were found between groups TH7 and TL7 (Figure S4C). The correlation analysis results showed that anthranilic acid (vitamin L1) had positive correlations with genistein, glycitein, and daidzein (Figure 6C).

At day 28, in the positive ion mode, seven differential metabolites, including N-(omega)-hydroxyarginine, choline, and 1-myristoyl-sn-glycero-3-phosphocholine, were found to have intergroup differences between groups T28 and TL28 (Figure S4D). The correlation analysis results showed positive correlations between taurine and N-acetyl-L-alanine, 1-myristoyl-sn-glycero-3-phosphocholine, and negative correlations between N-(omega)-hydroxyarginine and taurine, choline (Figure 6D). When comparing groups TL28 and TH28, 11 differential expressed metabolites were found in the negative ion mode (Figure S4E). The correlation analysis results showed negative correlations between 1,4-dihydroxybenzene and 3-guanidinopropanoate, and positive correlations between 1,4-dihydroxybenzene and dodecanoic acid, 12(R)-HETE, ethyl glucuronide, and hippuric acid. Additionally, hippuric acid showed a negative correlation with 3-guanidinopropanoate and positive correlations with 12(R)-HETE, ethyl glucuronide (Figure 6E). In the positive ion mode, 20 differential expressed metabolites were found (Figure S4F). The correlation analysis results showed negative correlations between anthranilic acid (vitamin L1) and 1-stearoyl-2-oleoyl-sn-glycerol 3-phosphocholine (SOPC), N4-acetylcytidine, cytosine, dimethylformamide, Lys-Gly. Conversely, anthranilic acid (vitamin L1) showed positive correlations with acetylcholine, equol, L-valine, 1-methylhistidine (Figure 6F).
KEGG pathway analysis of differential metabolites
The intergroup differential expressed metabolites between groups C7 and T7 were mainly enriched in the pathways of phenylalanine, tyrosine and tryptophan biosynthesis, arginine and proline metabolism, and melanogenesis (Figure 7A). The intergroup differential expressed metabolites between groups T7 and TL7 were mainly enriched in the pathways of aldosterone-regulated sodium reabsorption, prostate cancer, and cortisol synthesis and secretion (Figure 7B). No enriched pathways were found between groups TH7 and TL7. The intergroup differential expressed metabolites between groups C28 and T28 were mainly enriched in the pathways of pantothenate and coenzyme A (CoA) biosynthesis, beta-Alanine metabolism, and vitamin digestion and absorption (Figure 7C). The intergroup differential expressed metabolites between groups T28 and TL28 were mainly enriched in the pathways of bile secretion, ATP-binding cassette (ABC) transporters, and choline metabolism in cancer (Figure 7D). The intergroup differential expressed metabolites between groups TL28 and TH28 were mainly enriched in the pathways of central carbon metabolism in cancer, choline metabolism in cancer, and bile secretion (Figure 7E). Furthermore, we specifically analyzed the enrichment of differential metabolites among groups in the metabolic pathways (Figure S5). Compared to the control group, 1-methylhistidine was upregulated at day 7 and downregulated at day 28 in the experimental group (Figure 8A,8B). Compared to the experimental group, dodecanoic acid was downregulated in the Epacadostatlow group at both day 7 and day 28 (Figure 8C,8D). Compared to the Epacadostatlow group, 12(R)-HETE and hippuric acid were downregulated at both day 7 and day 28, while anthranilic acid (vitamin L1) was upregulated at day 7 and downregulated at day 28 in the Epacadostathigh group (Figure 8E,8F).


Discussion
Early-stage lung adenocarcinoma often lacks obvious symptoms, making accurate diagnosis challenging if based on clinical presentations alone. Early disruptions in cellular metabolism are hallmark features of cancer (26). The comprehensive application of metabolomics in substances such as lipids, amino acids, and organic acids has increased the potential for identifying useful new biomarkers for diagnosis and prognosis (27,28). In this study, a mouse model of early-stage lung adenocarcinoma was established, and UPLC-QTOF-MS analysis was used to analyze blood and urine samples extracted from mice. A total of 348 metabolites were identified, with organic acids and derivatives (16.954%) and lipids and lipid-like molecules (15.517%) being the two major classes of metabolites in early-stage lung adenocarcinoma mice. This finding is consistent with previous research results (29-31), indicating that these two classes of metabolites play important roles in the development of early-stage lung adenocarcinoma.
In terms of univariate statistical analysis, volcano plots and OPLS-DA analysis were used in this study to observe changes in differentially expressed metabolites and identify several metabolites with differential expression. These metabolite changes may be related to the occurrence and development of early-stage lung adenocarcinoma. For example, ethyl glucuronide, a metabolite related to ethanol metabolism, may be elevated in relation to alcohol consumption in lung cancer patients (32). Glyceric acid and myristic acid, two fatty acids, may be decreased due to the dependence of lung cancer cells on lipid metabolism (33). Hydrocortisone (cortisol), a hormone, may be elevated in relation to stress response in lung cancer patients (34). Anthranilic acid and N-acetylneuraminic acid, two aromatic metabolites, may be elevated in relation to inflammatory response (35,36).
Subsequently, the significantly different metabolites selected were subjected to cluster analysis and correlation analysis. The results of cluster analysis showed a certain clustering tendency in the expression patterns of metabolites among different samples. This indicates that specific groups of metabolites cluster together during the development of early-stage lung adenocarcinoma, and their expression patterns are similar. By further analyzing these clustered metabolites, we can identify some metabolic pathways associated with early-stage lung adenocarcinoma. Correlation analysis revealed positive or negative relationships between metabolites. These correlation relationships may reflect the mutual regulation of metabolic pathways during the occurrence and development of early-stage lung adenocarcinoma. Positively correlated metabolites may participate in common metabolic pathways or reaction chains, and their expression patterns are coordinated in early-stage lung adenocarcinoma. Negatively correlated metabolites may involve competitive metabolic pathways or inhibitory relationships. Among them, anthranilic acid (vitamin L1) under positive ion mode was found to be correlated with the expression levels of many metabolites in different groups. This suggests that anthranilic acid may be involved in metabolic regulation mechanisms during the occurrence and development of early-stage lung adenocarcinoma. Furthermore, in the subsequent KEGG enrichment analysis, a specific analysis of the Metabolic pathways revealed that anthranilic acid (vitamin L1) in the experimental group was upregulated on day 7 and downregulated on day 28 compared to the control group. Anthranilic acid is produced in the kynurenine pathway (KP) as a metabolite of kynurenine, which has been identified as an endogenous ligand for the human aryl hydrocarbon receptor (AHR) and a key factor in regulating immune response and cancer progression (35). In the tumor microenvironment, enzymes tryptophan-2,3-dioxygenase (TDO) and IDO1 are mainly responsible for activating the KP and are associated with anti-tumor immune suppression (37). Xiang et al. (38) found that IDO1 promotes cancer cell metastasis in gastric cancer, and the process involves changes in extracellular matrix components mediated by KP metabolites. Overactivation of the KP has also been observed in breast cancer and shown to be a key metabolic pathway contributing to immune escape (39). Furthermore, there is increasing evidence to suggest the potential therapeutic abilities of anthranilic acid as an anti-inflammatory and anticancer agent, with the design and synthesis of a series of anthranilic acid derivatives with certain anticancer properties (40,41). The results of this study demonstrate its involvement in the metabolic regulation process of lung adenocarcinoma.
In addition, this study also found that the differentially expressed metabolites between the C7 group and T7 group were mainly enriched in pathways such as phenylalanine, tyrosine and tryptophan biosynthesis, arginine and proline metabolism, and melanogenesis. This suggests that these pathways may also play important regulatory roles in the early development of lung adenocarcinoma. Similarly, the differentially expressed metabolites between the T7 group and TL7 group were mainly enriched in pathways such as aldosterone-regulated sodium reabsorption, prostate cancer, and cortisol synthesis and secretion, which may be related to the development and metabolic regulation of lung adenocarcinoma. Furthermore, compared to the Epacadostatlow group, the Epacadostathigh group showed upregulation of 1-methylhistidine expression on day 7 and downregulation on day 28. This seems to suggest a role for 1-methylhistidine in the treatment of lung adenocarcinoma with epacadostat as an IDO inhibitor. 1-Methylhistidine is a metabolite of amino acid metabolism associated with muscle protein degradation (42). In the field of cancer, research on 1-methylhistidine suggests its potential relationship with cancer development and metabolic regulation (43). Previous study has shown that cancer cachexia is associated with abnormal metabolism of 1-methylhistidine (44). As one of the markers of muscle protein degradation, 1-methylhistidine plays an important role in the development of cancer cachexia (45). Cancer cells have highly active metabolic states and require a large amount of amino acids to support their growth and division. To meet the energy and nitrogen source requirements of cancer cells, the body may increase protein degradation to release amino acids. This results in an increase in muscle protein degradation products such as 1-methylhistidine (45). Therefore, the level of 1-methylhistidine may be associated with muscle loss and the cachexia status of cancer. The correlation analysis results of this study showed a positive correlation between 1-methylhistidine and taurine, N-acetyl-L-alanine, and 1-myristoyl-sn-glycero-3-phosphocholine. This may indicate its important role in these metabolites or pathways involved in the regulation of lung adenocarcinoma metabolism.
Unfortunately, 12(R)-HETE and hippuric acid have been less studied in lung cancer, both in cell/mouse models and in lung cancer patients. 12(R)-HETE, is a metabolite derived from arachidonic acid. 12(R)-HETE is formed by the metabolism of arachidonic acid by enzymes such as 12- and 15-lipoxygenase in human platelets and polymorphonuclear leukocytes and has several biological activities that can be assessed in models of inflammation. Hippuric acid has been described as the intestinal microbial mammalian co-metabolite of benzoic acid. This means that both gut microbes and mammalian cells (especially liver cells) are involved in its formation. However, no studies have found its role in lung adenocarcinoma. Our study is the first to propose the effect of IDO inhibitors on 12(R)-HETE and hippuric acid, and its mechanism needs further study.
Furthermore, it should be noted that while human samples have been extensively studied in previous research, the mouse model provides us with a unique opportunity to explore the early stages of the disease, which is difficult to achieve with human samples. The use of murine lung tumor models, while not directly translatable to human disease, offers several advantages. Firstly, it allows for controlled studies in a genetically homogeneous population, eliminating many confounding factors present in human studies. Secondly, mouse models provide an opportunity to explore tumor development and progression in real-time, enabling a more comprehensive understanding of the disease. However, it is essential to note that findings from this study should be interpreted with caution when translating to human lung adenocarcinoma. We acknowledge this as a limitation, as mouse models may not fully reflect all nuances of human disease and further studies using human samples will be needed to validate our findings.
Conclusions
This study reveals metabolic alterations associated with early-stage lung adenocarcinoma in mice. The identified differentially expressed metabolites and enriched pathways provide potential targets for further investigation and emphasize the importance of metabolomics analysis in understanding the underlying mechanisms of lung adenocarcinoma. The discovery of these differential metabolites further highlights the significance of metabolic dysregulation in the early stages of lung adenocarcinoma. The changes in ethyl glucuronide and glyceric acid may be related to the modulation of metabolic pathways in tumor cells, while the upregulation of anthranilic acid may be associated with the body’s metabolic response to lung adenocarcinoma. However, further research is needed to elucidate the specific functions of these differential metabolites and their relationship with the development of lung adenocarcinoma.
This study provides clues for further investigation into the early diagnosis and treatment of lung adenocarcinoma. By gaining a deeper understanding of the biological significance of these differential metabolites, we may discover new biomarkers or therapeutic targets, providing better strategies for personalized treatment and management of lung adenocarcinoma patients.
Acknowledgments
Funding: This study was funded by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1236/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1236/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1236/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1236/coif). L.M. received funding from the Project of Hunan Provincial Health Commission (No. 20200247), the Hunan Province Clinical Medical Technology Innovation Guidance Project (No. 2020SK51825), the Hunan Provincial Natural Science Foundation (No. 2021JJ30618), the Hunan Province Degree and Graduate Teaching Reform Research Project (No. 2022JGYB172), and the Ministry of Education’s Industry School Cooperation Collaborative Education Project (No. 220601867221601). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. 2019LL1021001) granted by the Ethics Committee of the First Affiliated Hospital, University of South China, in compliance with national or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Isaksson S, Jönsson P, Monsef N, et al. CA 19-9 and CA 125 as potential predictors of disease recurrence in resectable lung adenocarcinoma. PLoS One 2017;12:e0186284. [Crossref] [PubMed]
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Su L, Zhao J, Su H, et al. CircRNAs in Lung Adenocarcinoma: Diagnosis and Therapy. Curr Gene Ther 2022;22:15-22. [PubMed]
- Spranger S, Koblish HK, Horton B, et al. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. J Immunother Cancer 2014;2:3. [Crossref] [PubMed]
- Pertovaara M, Hasan T, Raitala A, et al. Indoleamine 2,3-dioxygenase activity is increased in patients with systemic lupus erythematosus and predicts disease activation in the sunny season. Clin Exp Immunol 2007;150:274-8. [Crossref] [PubMed]
- Mellor A. Indoleamine 2,3 dioxygenase and regulation of T cell immunity. Biochem Biophys Res Commun 2005;338:20-4. [Crossref] [PubMed]
- Tang K, Wu YH, Song Y, et al. Indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors in clinical trials for cancer immunotherapy. J Hematol Oncol 2021;14:68. [Crossref] [PubMed]
- Azimnasab-Sorkhabi P, Soltani-Asl M, Yoshinaga TT, et al. Indoleamine-2,3 dioxygenase: a fate-changer of the tumor microenvironment. Mol Biol Rep 2023;50:6133-45. [Crossref] [PubMed]
- Kelly CM, Qin LX, Whiting KA, et al. A Phase II Study of Epacadostat and Pembrolizumab in Patients with Advanced Sarcoma. Clin Cancer Res 2023;29:2043-51. [Crossref] [PubMed]
- Powderly JD, Klempner SJ, Naing A, et al. Epacadostat Plus Pembrolizumab and Chemotherapy for Advanced Solid Tumors: Results from the Phase I/II ECHO-207/KEYNOTE-723 Study. Oncologist 2022;27:905-e848. [Crossref] [PubMed]
- Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, et al. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol 2022;20:143-60. [Crossref] [PubMed]
- Suman S, Sharma RK, Kumar V, et al. Metabolic fingerprinting in breast cancer stages through (1)H NMR spectroscopy-based metabolomic analysis of plasma. J Pharm Biomed Anal 2018;160:38-45. [Crossref] [PubMed]
- Guan W, Zhou M, Hampton CY, et al. Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinformatics 2009;10:259. [Crossref] [PubMed]
- Farrokhi Yekta R, Rezaei Tavirani M, Arefi Oskouie A, et al. Serum-based metabolic alterations in patients with papillary thyroid carcinoma unveiled by non-targeted 1H-NMR metabolomics approach. Iran J Basic Med Sci 2018;21:1140-7. [PubMed]
- Gao Y, Chen Y, Yue X, et al. Development of simultaneous targeted metabolite quantification and untargeted metabolomics strategy using dual-column liquid chromatography coupled with tandem mass spectrometry. Anal Chim Acta 2018;1037:369-79. [Crossref] [PubMed]
- Schmahl MJ, Regan DP, Rivers AC, et al. NMR-based metabolic profiling of urine, serum, fecal, and pancreatic tissue samples from the Ptf1a-Cre; LSL-KrasG12D transgenic mouse model of pancreatic cancer. PLoS One 2018;13:e0200658. [Crossref] [PubMed]
- Wu H, Xue R, Tang Z, et al. Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem 2010;396:1385-95. [Crossref] [PubMed]
- Fan L, Zhang W, Yin M, et al. Identification of metabolic biomarkers to diagnose epithelial ovarian cancer using a UPLC/QTOF/MS platform. Acta Oncol 2012;51:473-9. [Crossref] [PubMed]
- Hu JM, Sun HT. Serum proton NMR metabolomics analysis of human lung cancer following microwave ablation. Radiat Oncol 2018;13:40. [Crossref] [PubMed]
- Carrola J, Rocha CM, Barros AS, et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res 2011;10:221-30. [Crossref] [PubMed]
- Kawamoto H, Hara H, Araya J, et al. Prostaglandin E-Major Urinary Metabolite (PGE-MUM) as a Tumor Marker for Lung Adenocarcinoma. Cancers (Basel) 2019;11:768. [Crossref] [PubMed]
- Han X, Luo R, Wang L, et al. Potential predictive value of serum targeted metabolites and concurrently mutated genes for EGFR-TKI therapeutic efficacy in lung adenocarcinoma patients with EGFR sensitizing mutations. Am J Cancer Res 2020;10:4266-86. [PubMed]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21:297-308. [Crossref] [PubMed]
- Fahrmann JF, Kim K, DeFelice BC, et al. Investigation of metabolomic blood biomarkers for detection of adenocarcinoma lung cancer. Cancer Epidemiol Biomarkers Prev 2015;24:1716-23. [Crossref] [PubMed]
- Wikoff WR, Grapov D, Fahrmann JF, et al. Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev Res (Phila) 2015;8:410-8. [Crossref] [PubMed]
- Yang Q, Luo J, Xu H, et al. Metabolomic investigation of urinary extracellular vesicles for early detection and screening of lung cancer. J Nanobiotechnology 2023;21:153. [Crossref] [PubMed]
- Xiang Y, Zhao Q, Wu Y, et al. Serum Metabolomics Profiling Reveals Metabolic Alterations Prior to a Diagnosis with Non-Small Cell Lung Cancer among Chinese Community Residents: A Prospective Nested Case-Control Study. Metabolites 2022;12:906. [Crossref] [PubMed]
- Deng H, Li W. Monoacylglycerol lipase inhibitors: modulators for lipid metabolism in cancer malignancy, neurological and metabolic disorders. Acta Pharm Sin B 2020;10:582-602. [Crossref] [PubMed]
- Troche JR, Mayne ST, Freedman ND, et al. Alcohol Consumption-Related Metabolites in Relation to Colorectal Cancer and Adenoma: Two Case-Control Studies Using Serum Biomarkers. PLoS One 2016;11:e0150962. [Crossref] [PubMed]
- Yan L, Rust BM, Sundaram S, et al. Alteration in Plasma Metabolome in High-Fat Diet-Fed Monocyte Chemotactic Protein-1 Knockout Mice Bearing Pulmonary Metastases of Lewis Lung Carcinoma. Nutr Metab Insights 2022;15:11786388221111126. [Crossref] [PubMed]
- Yang H, Xia L, Chen J, et al. Stress-glucocorticoid-TSC22D3 axis compromises therapy-induced antitumor immunity. Nat Med 2019;25:1428-41. [Crossref] [PubMed]
- Opitz CA, Litzenburger UM, Sahm F, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011;478:197-203. [Crossref] [PubMed]
- Chou HH, Hayakawa T, Diaz S, et al. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci U S A 2002;99:11736-41. [Crossref] [PubMed]
- Prendergast GC, Malachowski WP, DuHadaway JB, et al. Discovery of IDO1 Inhibitors: From Bench to Bedside. Cancer Res 2017;77:6795-811. [Crossref] [PubMed]
- Xiang Z, Li J, Song S, et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res 2019;38:314. [Crossref] [PubMed]
- Heng B, Lim CK, Lovejoy DB, et al. Understanding the role of the kynurenine pathway in human breast cancer immunobiology. Oncotarget 2016;7:6506-20. [Crossref] [PubMed]
- Şenol H, Çağman Z, Gençoğlu Katmerlikaya T, et al. New Anthranilic Acid Hydrazones as Fenamate Isosteres: Synthesis, Characterization, Molecular Docking, Dynamics & in Silico ADME, in Vitro Anti-Inflammatory and Anticancer Activity Studies. Chem Biodivers 2023;20:e202300773. [Crossref] [PubMed]
- Abdelkader DH, Elekhnawy E, Negm WA, et al. Insight into Fucoidan-Based PEGylated PLGA Nanoparticles Encapsulating Methyl Anthranilic Acid: In Vitro Evaluation and In Vivo Anti-Inflammatory Study. Mar Drugs 2022;20:694. [Crossref] [PubMed]
- Davydova E, Shimazu T, Schuhmacher MK, et al. The methyltransferase METTL9 mediates pervasive 1-methylhistidine modification in mammalian proteomes. Nat Commun 2021;12:891. [Crossref] [PubMed]
- Cross AJ, Major JM, Rothman N, et al. Urinary 1-methylhistidine and 3-methylhistidine, meat intake, and colorectal adenoma risk. Eur J Cancer Prev 2014;23:385-90. [Crossref] [PubMed]
- Ubhi BK, Cheng KK, Dong J, et al. Targeted metabolomics identifies perturbations in amino acid metabolism that sub-classify patients with COPD. Mol Biosyst 2012;8:3125-33. [Crossref] [PubMed]
- Baracos VE, Martin L, Korc M, et al. Cancer-associated cachexia. Nat Rev Dis Primers 2018;4:17105. [Crossref] [PubMed]