
Disulfidptosis and ferroptosis related genes predict prognosis and personalize treatment for hepatocellular carcinoma
Highlight box
Key findings
• The disulfidptosis and ferroptosis (DRG-FRG) genes signature effectively categorized patients into two distinct categories based on their risk levels, emphasizing significant biological pathways, therapeutic targets, immunological landscape, and checkpoint molecules.
What is known and what is new?
• The manuscript explores DRG-FRG signature risk score, clinical characteristics, protein node significance, and immune infiltration in hepatocellular carcinoma (HCC) progression.
• The manuscript explores molecular mechanisms, identifies potential drugs, and analyzes drug sensitivity.
What is the implication, and what should change now?
• The findings highlight the importance of DRG-FRG genes in HCC, as well as their interaction with diverse biological processes and pathways, and prospective small molecule medicines offer novel therapeutic paths for HCC treatment.
• Given the manuscript’s findings on HCC-related small molecule medications, immunological checkpoint expression profiles, immune checkpoint blockade therapy should be investigated.
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and a leading cause of illness and death around the world (1). The incidence of HCC varies depending on the geographic region, with the highest rates found in Asia and sub-Saharan Africa. The most important risk factors for developing HCC are chronic hepatitis B and C infections, as well as alcohol consumption and exposure to aflatoxin. Unfortunately, the mortality rate for HCC is also high, and patients with advanced HCC have a poor prognosis. While there have been notable strides in the treatment of HCC through immunotherapy, targeted therapy, and liquid biopsy, the survival rate for HCC patients is still comparatively low (2). This is largely due to factors such as the aggressiveness of the disease, late detection, and the presence of other medical conditions. In light of this, it is imperative that more effective treatments are developed and implemented to improve the survival rates of HCC patients. Multigene signatures are important in cancer research, they help evaluate the outcomes of treatments and identify high-risk patients for earlier intervention. With the potential to revolutionize cancer research and treatment, multigene signatures provide a promising avenue for medical practitioners and researchers. In this context, our research seeks to develop a signature that combines disulfidptosis and ferroptosis (DRG-FRG), aiming to forecast the overall survival and treatment options of HCC patients.
Disulfidptosis is a novel type of cell death that occurs when disulfides, such as cystine, accumulate in cancer cells during glucose starvation. This abnormal buildup of intracellular disulfides in SLC7A11 cells results in a type of cell death which is different from both apoptosis and ferroptosis (3). The consequence of this is the collapse of the actin cytoskeleton, leading to the loss of cell shape and integrity (4). This phenomenon may offer a potential target for metabolic cancer therapy, as it can be induced by glucose transporter inhibitors or other agents that deplete NADPH or increase disulfide stress (4,5). Additionally, other influential studies have suggested that disulfidptosis may have implications for tumor immunity, as the cell death signal may activate tumor-specific T cells and enhance the anti-tumor immune response (6,7). SLC7A11 overexpression can promote tumor growth and suppress ferroptosis in some cancers, but may also increase cancer cell dependence on glucose and glutamine (8).
The process of ferroptosis involves the build-up of iron-dependent reactive oxygen species as well as lipid peroxidation, this programmed cell death differs from apoptosis and necrosis (9,10). Ferroptosis has been shown to have both tumor-suppressive and tumor-promoting effects in HCC, depending on the context and the signaling pathways involved (11). Understanding the role of ferroptosis in HCC development and progression is essential for developing effective therapies. Inhibition of ferroptosis has been shown to promote the growth of HCC, while induction of ferroptosis inhibits HCC cell growth and could enhance the efficacy of chemotherapy. Targeting ferroptosis could be a promising therapeutic strategy for HCC patients (12).
DRG-FRG are both forms of cell death that are thought to be involved in cancer progression and treatment. The DRG-FRG signature is expected to be a useful tool for medical practitioners and researchers in the field of oncology to evaluate the outcomes of treatments and identify high-risk HCC patients for earlier intervention. However, there have been few studies that systematically explore the relationship between DRG-FRG and HCC.
Our research focused intensively on the bioinformatics analyses to explore the expression spectrum and prognostic significance of DRG-FRG in HCC. We crafted and validated a predictive feature rooted in 11 DRG-FRGs, which showcased its prowess in predicting the prognosis of HCC patients. Notably, a link was discerned between HCC’s prognostic indicators and the immune landscape, laying a foundational theory for the adoption of immune checkpoint therapies. On top of that, we pinpointed ten small molecule drugs that hold promise for patient benefits. We present this article in accordance with the STROBE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1594/rc).
Methods
Collecting data and identifying differentially expressed DRG-FRG
In our study, we undertook a detailed analysis of 424 hepatic tissue samples, which included 50 normal liver tissue samples and 374 HCC tissue samples. This analysis involved evaluating messenger RNA (mRNA) expression levels and incorporating pertinent clinical data sourced from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov). From earlier thematic studies, we retrieved 23 disulfideptosis genes (3) and 79 immune checkpoint genes (13). Ferroptosis genes were obtained from the FerrDb V2 (http://www.zhounan.org/ferrdb/current/). After meticulously merging and refining three relevant dataset files, which were all sourced from the FerrDb V2 database (tables available at https://cdn.amegroups.cn/static/public/tcr-23-1594-1.xlsx; https://cdn.amegroups.cn/static/public/tcr-23-1594-2.xlsx; https://cdn.amegroups.cn/static/public/tcr-23-1594-3.xlsx), we pinpointed a total of 259 ferroptosis genes. We extracted the mRNA expression patterns of both disulfideptosis and ferroptosis from the TCGA dataset. Subsequently, the limma package was employed to perform correlation analysis in order to identify ferroptosis-associated genes and their corresponding mRNA expression levels that exhibit significant associations with disulfideptosis. The selection criteria for these genes were as follows: the correlation coefficient should be >0.3 (indicating positive correlation) or <−0.3 (indicating negative correlation), and the P<0.05. Genes meeting these criteria were classified as DRG-FRG genes. The process to identify differentially expressed DRG-FRG genes between normal liver tissue and HCC tissue samples adhered to specific criteria: |logfold change (logFC)| of >1 and false discovery rate (FDR) of <0.05, once more utilizing the limma package in R software. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
To assess the prognostic value of DRG-FRG, we initiated a univariate Cox regression analysis. Building on this, least absolute shrinkage and selection operator (LASSO) penalized Cox regression analysis was utilized, with the aid of the glmnet R package, to craft the prognostic risk model. Risk scores were determined based on the formula outlined in the literature (14). This led to the stratification of the HCC group into low-risk and high-risk categories using median risk scores. For a deeper understanding of patient outcomes, the Kaplan-Meier curve was used to analyze the survival rates of these two distinct patient groups. To further gauge the predictive accuracy of the risk model, a time-related receiver operating characteristic (ROC) analysis was executed, leveraging the survival ROC package in R software. Lastly, a validation set, comprising 70% of the initial model, was randomly chosen to evaluate the model’s prognostic efficacy.
A nomogram integrating the DRG-FRG risk score and clinical variables enabled individualized outcome prediction for HCC
Our analysis primarily delved into the relationship between the risk score of the DRG-FRG signature and clinical characteristics in HCC patients. Beyond this, univariate and multivariate Cox regressions were executed, incorporating various clinical covariates, to ascertain if the DRG-FRG risk score could stand as an independent prognostic marker for HCC. To provide a comprehensive predictive tool, a prognostic nomogram was formulated, integrating both clinical variables and the DRG-FRG signature risk score, aiming to forecast the 1-, 3-, and 5-year overall survival rates for HCC patients. To ensure its reliability, the nomogram’s predictive accuracy was assessed using the concordance index (C-index) and calibration analysis.
Biological enrichment studies and analyses of protein-protein interaction (PPI)
Gene Ontology (GO) analysis, encompassing biological process (BP), cellular component (CC), molecular function (MF), as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, were performed using the ClusterProfiler software. A term was deemed significantly enriched when both its FDR and P<0.05. For further insights, differentially expressed DRG-FRG were inputted into the STRING database (version 11.0; https://cn.string-db.org/) to obtain PPI data. Only PPI pairs boasting a combined score above 0.4 were chosen, leading to the creation and visualization of the PPI network via Cytoscape software (version 3.9.1) (15). Utilizing CytoHubba, a plugin within Cytoscape, we gauged the significance of individual protein nodes (16). In our methodology, hub genes were pinpointed using the Maximal Clique Centrality (MCC) approach. They were then ranked by their MCC scores, and the top 10 hub genes were earmarked and presented.
Gene set enrichment analysis (GSEA)
We employed GSEA to pinpoint the biological pathways that differentiate high-risk from low-risk groups. GSEA facilitates the identification of genes that demonstrate coordinated up- or down-regulation within predefined sets, thereby spotlighting the crucial biological pathways in play. We set a significance threshold at P<0.05 to ascertain statistical relevance. Through the integration of GSEA in our study, our aim is to offer a comprehensive exploration of the biological processes and pathways pivotal to risk differentiation. This approach not only enhances our understanding of the molecular intricacies but also sets the stage for future specialized investigations and potential therapeutic interventions.
Screening for potential small molecule drugs
We uploaded the 11 differentially expressed DRG-FRG signatures into Enrichr (https://maayanlab.cloud/Enrichr/). Following this, we employed the Drug Signatures Database (DSigDB; http://tanlab.ucdenver.edu/DSigDB) to pinpoint potential small molecule drugs (17). Only compounds with an adjusted P<0.05 were deemed to have a statistically meaningful correlation with the uploaded genes.
The analysis of drug sensitivity
To delve into the differences in drug sensitivity between the high-risk and low-risk groups, we turned to the Genomics of Drug Sensitivity in Cancer (GDSC; https://www.cancerrxgene.org/) database. Utilizing this resource, we conducted an analysis of the drug’s half-maximal inhibitory concentration (IC50) using the pRRophetic package, aiming to predict drug responsiveness. If the P<0.05, it is deemed to have statistical significance.
The analysis of immune cell infiltration base on DRG-FRG genes
Recent studies have underscored that immune infiltration of tumor cells plays a pivotal role in cancer progression and outcomes (17,18). In light of this, leveraging the B-cell specific long non-coding RNA (lncRNA) signature, we employed a suite of algorithms, namely QUANTISEQ (http://icbi.at/quantiseq), CIBERSORT (https://cibersortx.stanford.edu), XCELL (https://comphealth.ucsf.edu/app/xcell), CIBERSORT abs.mode (https://cibersortx.stanford.edu), EPIC (http://epic.gfellerlab.org), TIMER (https://cistrome.shinyapps.io/timer), and MCP-counter (https://github.com/ebecht/MCPcounter), to gauge the extent of immune cell infiltration across high- and low-risk groups. Additionally, we delved into the expression profiles of several immunological checkpoints pertinent to HCC, such as PDCD1, LAG3, HAVCR2, TIGIT, CTLA4, TIM-3, GITR, CD27, CD28, and OX40, aiming to forecast the efficacy of immune checkpoint blockade treatments. The TIMER2 database (http://timer.cistrome.org) was instrumental in shedding light on the interplay between immune cells and 11 specific DRG-FRG, thereby enriching our comprehension of the functional significance of DRG-FRG in HCC.
Statistics analysis
We conducted all statistical analyses using the R software (version 4.2.3). A threshold of two-sided P<0.05 was set to determine statistical significance.
Results
Construction of a gene dataset for DRG-FRG
After merging the HCC-associated mRNA dataset from TCGA with the 23 disulfidptosis genes, it was evident that expression data were available for all these genes across both the normal and HCC groups (table available at https://cdn.amegroups.cn/static/public/tcr-23-1594-4.xlsx). In a parallel manner, when integrating the ferroptosis genes with the HCC-associated mRNA dataset, 240 out of the 259 ferroptosis genes showcased expression data for both groups (table available at https://cdn.amegroups.cn/static/public/tcr-23-1594-5.xlsx). The correlation between DRG-FRG, grounded on our selection criteria, is detailed in table available at https://cdn.amegroups.cn/static/public/tcr-23-1594-6.xlsx. Streamlining our datasets, we amalgamated the disulfidptosis-related gene expression data with that of the ferroptosis-related genes, resulting in the DRG-FRG genes expression matrix (table available at https://cdn.amegroups.cn/static/public/tcr-23-1594-7.xlsx).
Identification and validation of differential expression DRG-FRG genes
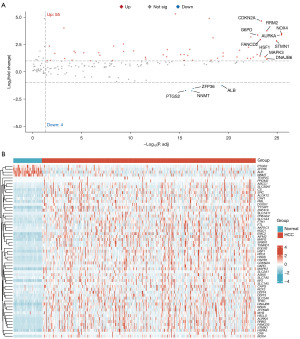
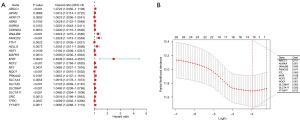
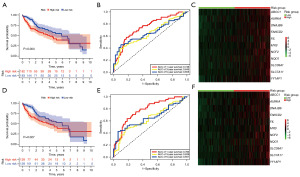
In the differential expression analysis of the TCGA-HCC dataset, 59 DRG-FRG genes exhibited differential expression when juxtaposed with normal liver tissues: four were downregulated, while 55 saw upregulation (Figure 1A,1B). To ascertain their prognostic relevance with HCC, we conducted a univariate Cox regression analysis on these altered DRG-FRG genes. This revealed that 24 genes held prognostic significance, with all being earmarked as genes indicating poor prognosis for HCC, each having a hazard ratio exceeding 1 (Figure 2A). Subsequently, a LASSO Cox regression was executed to craft a prognostic signature from the aforementioned 24 DRG-FRG genes. This analysis pinpointed 11 DRG-FRG genes (ABCC1, AURKA, DNAJB6, FANCD2, FTL, MYB, NCF2, NQO1, SLC38A1, SLC7A11, and YY1AP1) for the risk model (Figure 2B). The risk score was derived using the formula detailed in the methodology, factoring in the coefficients linked to the 11 DRG-FRG genes. HCC patients were then bifurcated into high-risk and low-risk factions based on median risk scores. Interestingly, the high-risk faction registered a markedly elevated mortality rate in contrast to the low-risk group (P=0.004) (Figure 3A). The time-dependent ROC analysis showcased the risk score model’s accuracy rates of 0.746, 0.645, and 0.655 for 1-, 3-, and 5-year survival predictions respectively (Figure 3B). Heatmap representations further highlighted the pronounced expression of the 11 DRG-FRG genes within the high-risk group (Figure 3C). For validation purposes, we earmarked 70% of the original dataset as the test set, which mirrored the findings of the primary model (Figure 3D-3F).
The prognosis of HCC was determined independently by the signature based on the DRG-FRG.
To evaluate the signatures’ capability as standalone prognostic indicators, we undertook both univariable and multivariable Cox analyses, factoring in age, gender, tumor grade, pathologic stage, and risk score. The univariate analysis underscored a notable correlation between the risk score and pathologic stage in relation to HCC patient survival (P=0.007 and P<0.001, respectively) (Figure 4A). Furthermore, the multivariate analysis affirmed the significant impact of both the risk score and pathologic stage on prognosis (P=0.022 and P<0.001) (Figure 4B). These insights suggest that the DRG-FRG-based signature holds promise as a reliable prognostic tool for HCC patients.

The correlation between the DRG-FRG-based signature and clinical characteristics
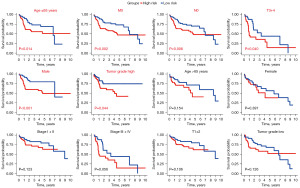
We probed the potential involvement of the prognostic signature in patients’ development and progression using the chi-squared test. As illustrated in Figure 5A,5B, there were pronounced differences between the two groups concerning pathologic stage (P=0.013), T stage (P=0.016), and tumor grade (P<0.001). However, factors like age, M stage, gender, and N stage did not exhibit significant variations (P>0.05). Subsequently, we categorized the signature based on diverse subgroups to ascertain its prognostic merit (Figure 6). Our analysis revealed that the DRG-FRG-based signature possessed a robust predictive capacity for subgroups such as age ≤65 years (P=0.014), high tumor grade (P=0.044), M0 (P=0.002), T3–T4 stage (P=0.040), N0 (P=0.008), and male (P<0.001). On the flip side, its predictive efficacy was limited in subgroups like age >65 years, female, pathologic stage, high tumor grade, and T1–T2 stage (P>0.05).

Construction of a nomogram for evaluating prognosis
We visually assessed an individual’s survival probability using a nomogram that integrated various prognostic factors. This nomogram, designed to further predict the survival outcomes of HCC patients, incorporated elements like tumor grade, age, gender, pathologic stage, and risk score. As depicted in Figure 7A, the nomogram facilitated the precise estimation of the 1-, 3-, and 5-year survival rates for HCC patients. For instance, the prognostic outcome for the 20th patient was visually represented, showcasing a cumulative score of 64.4. This translated to survival rates of 91.1% at 1 year, 79.9% at 3 years, and 70.4% at 5 years. The calibration curve in Figure 7B highlighted that the actual patient survival closely mirrored the nomogram’s predictions. Moreover, with a C-index of 0.785, the nomogram’s predictive prowess was affirmed.
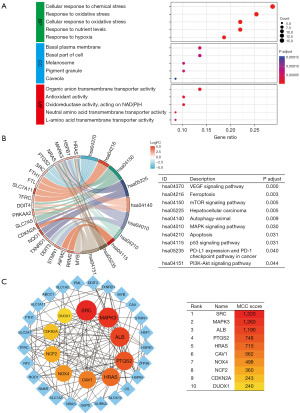
Biological enrichment analyses and PPI analyses
In the BP analysis, 59 DRG-FRG genes were prominently associated with cellular responses to chemical stress, oxidative stress, nutrient levels, and hypoxia. The CC analysis highlighted enrichment in areas such as the basal plasma membrane, basal part of the cell, melanosome, pigment granule, and caveola. The MF analysis revealed that these 59 DRG-FRG genes predominantly participated in functions like organic anion transmembrane transporter activity, antioxidant activity, oxidoreductase activity, and both neutral and L-amino acid transmembrane transporter activities (Figure 8A). KEGG analysis identified key signaling pathways in which these genes played a role, including VEGF, ferroptosis, mTOR, HCC, autophagy-animal, MAPK, apoptosis, p53, PD-L1 expression, PD-1 checkpoint pathway in cancer, and PI3K-Akt (Figure 8B). Utilizing the STRING database, a PPI network was constructed for the 35 differentially expressed DRG-FRG genes. All these genes exhibited high expression levels. This PPI network was further refined using R software. The top 10 hub genes, identified through the MCC algorithm of the cytoHubba plugin, included SRC, MAPK3, ALB, PTGS2, HRAS, CAV1, NOX4, NCF2, CDKN2A, and DUOX1. These genes were ranked based on their scores and depicted with varying color intensities (Figure 8C).
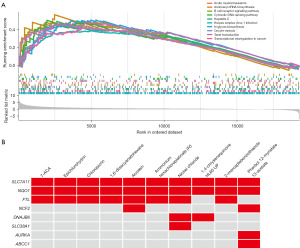
GSEA for DRG-FRG-based signature
To delve deeper into the molecular mechanisms that underpin the DRG-FRG-based signature, we employed GSEA. The insights gleaned from GSEA highlighted several pivotal biological pathways. Specifically, pathways such as aminoacyl-tRNA biosynthesis, B cell receptor signaling pathway, cytosolic DNA-sensing pathway, hepatitis C, herpes simplex virus 1 infection, N-glycan biosynthesis, oocyte meiosis, taste transduction, and transcriptional mis-regulation in cancer were found to be significantly enriched in the high-risk group (Figure 9A).
Screening of small molecule drugs based on 11 DRG-FRG genes
Utilizing the DsigDB database, we identified potential small molecule drugs associated with the 11 DRG-FRG genes. As depicted in Figure 9B, the genetic variations most pertinent to the top ten small molecule drugs are presented, ranked by the adjusted P value magnitude. Notably, our analysis underscored a pronounced association between the NQO1 and SLC7A11 genes with these ten small molecule drugs, followed by the FTL gene, which exhibited a distinct correlation with eight out of the ten small molecule drugs. Unfortunately, there is no evidence to suggest that FANCD2, MYB, and YY1AP1 genes have any association with the top ten small molecule drugs, and these three genes are not even listed in Figure 9B.
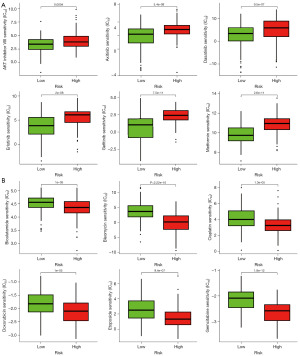
Correlational analysis of 11 DRG-FRG genes and drug sensitivity
Based on the data sourced from the GDSC database, the IC50 values for AKT inhibitor VIII, axitinib, dasatinib, erlotinib, gefitinib, and metformin were observed to be significantly elevated in the high-risk group compared to the low-risk group. This trend underscores a heightened drug resistance in high-risk patients (refer to Figure 10A). Conversely, the IC50 values for bicalutamide, bleomycin, cisplatin, doxorubicin, etoposide, and gemcitabine were markedly increased in the low-risk group, indicating a greater drug sensitivity in the high-risk cohort (Figure 10B).
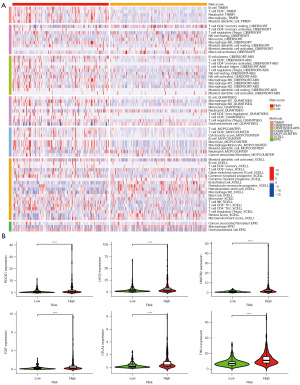
Examination of immune infiltration levels using the DRG-FRG-based signature
The heatmap depicted in Figure 11A elucidates the interplay between immune infiltration and the DRG-FRG-based signature. Notably, variations in the expression of immune-infiltrating cells were discerned between the high- and low-risk score groups. Recognizing the pivotal role of checkpoint inhibitor treatments, we honed in on the correlation between HCC-associated risk scores and key immune checkpoints, namely TIM-3, LAG3, PDCD1, TIGIT, HAVCR2, and CTLA4. Our findings highlight distinct expression differences of these immune checkpoints across the patient cohort, with the high-risk group manifesting markedly elevated levels, as showcased in Figure 11B.
Analysis of correlation between DRG-FRG genes and immune cells from the TIMER database
Utilizing the TIMER database, we probed the relationship between immune cells and the 11 prognostic DRG-FRG genes. Our analysis revealed that several genes, specifically ABCC1, DNAJB6, MYB, NCF2, NQO1, SLC7A11, and SLC38A1, exhibited a negative correlation with purity. Simultaneously, these genes manifested a positive association with B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. In contrast, AURKA, FANCD2, and YY1AP1 displayed a positive correlation with both purity and the aforementioned immune cells. FTL, on the other hand, was negatively correlated with purity and all the previously mentioned immune cells, with the exception of B cells (refer to Figures S1,S2).
Discussion
Apoptosis, autophagy, necroptosis, ferroptosis, and copperptosis represent prominent instances of programmed cell death, alongside the recently discovered disulfidptosis (4). This discovery provides new perspectives and avenues for in-depth research on the formation and progression of cancer, while also holding critical potential for advancing cancer treatment.
The present study aimed to develop a gene signature model utilizing disulfideptosis and ferroptosis related genes for the purpose of prognostic prediction and treatment selection in HCC. The collaborative examination of multiple genes offers researchers novel insights and methodologies to investigate the causation, progression, and therapeutic interventions of diseases.
For example, Wang et al. performed an examination of gene expression data acquired from the Gene Expression Omnibus. The primary aim of their investigation was to ascertain hub genes that are associated with the development and prognosis of HCC. Significantly, the researchers identified FCN3 and FOXO1 as pivotal genes that demonstrate promise as biomarkers for the early detection of HCC and as prognostic indicators for overall patient survival (19). Similarly, the recent genomic profiling of HCC tumors has unveiled a high occurrence of mutations in genes such as TERT, TP53, and CTNNB1. These mutations play a crucial role in identifying core deregulated pathways, defining molecular subtypes, and presenting potential therapeutic targets and prognostic biomarkers (20).
This study utilized LASSO regression analysis to screen genes with high prognostic value established a risk score model base on these genes. This model demonstrated significant efficacy in predicting overall survival among HCC patients, while also serving as an independent predictor, apart from other clinical variables, such as pathological stage. Ye et al. carried out an in-depth analysis of the autophagy-associated genes in HCC and developed a prognostic model centered on these genes. Their study highlighted the crucial role played by autophagy in the progression and prognosis of HCC (21). Wang et al. conducted a study in which they identified pivotal immune-related genes in patients diagnosed with HCC and subsequently employed these genes to develop a prognostic model. The efficacy of this model, capable of autonomously predicting patient outcomes and reflecting the immune status of the tumor, was further verified using an independent database (22). The aforementioned studies, along with the ongoing research, significantly contribute to the advancement of understanding prognosis in HCC. Consequently, the incorporation of the 11-gene DRG-FRG signature as a means to extract dependable prognostic biomarkers from cancer omics data offers an additional approach. This has the potential to facilitate personalized risk assessment and inform clinical decision-making in the context of HCC.
MYB is a gene associated with poor prognosis in HCC patients, its’ HR (derived from univariate Cox regression analysis) and coefficient (derived from LASSO Cox regression analysis) are significantly higher than all others, which suggests that MYB has a more significant impact on the prognosis of HCC patients compared to other genes. The findings from our study are in line with previous literature (23). Furthermore, the survival curves for low- and high-risk groups intersected and reversed after about 7 years according to Figure 3A,3D. A possible explanation for this result is that the number of survivors and the survival rate for both groups become very low after 7 years. However, the 5-year survival rate for HCC is only around 10% worldwide (1), and the rate is 10.96% (40 out of 365) in our study. Therefore, the 5-year survival rate of HCC patients remains a crucial research topic. From Figure 3A,3D, we can see that in the first 5 years, the survival rate of the low-risk group patients (blue curve) is significantly higher than that of the high-risk group patients (red curve). Therefore, our conclusion is reliable.
This study has successfully identified a distinct DRG-FRG signature consisting of 11 genes that can effectively forecast the prognosis and chemotherapy responsiveness in patients with HCC. Notably, recent mechanistic and clinical investigations have established the involvement of several genes within this signature, such as ABCC1, SLC7A11, AURKA, and NQO1, in the progression of HCC and the development of resistance to treatment. The overexpression of ABCC1 actively removes drug conjugates from cellular compartments, fostering the progression of chemotherapy resistance in HCC, leading to poor prognosis in patients (24-26). The downregulation or inhibition of ABCC1 renders resistant HCC cells more susceptible to the cytotoxic effects of doxorubicin and other chemotherapeutic agents, highlighting its potential as both a prognostic indicator and a therapeutic target (27,28). SLC7A11 serves as a crucial transporter involved in the synthesis of glutathione, thereby playing a pivotal role in maintaining redox homeostasis to counteract ferroptosis. The upregulation of SLC7A11 in HCC is closely linked to venous infiltration, advanced tumor stage, and unfavorable survival outcomes (29,30). SOCS2 interacts with and promotes ubiquitination-mediated degradation of SLC7A11, thereby inducing iron-dependent ferroptosis cell death and increasing radiosensitivity of HCC (31). In preclinical studies, the inhibition of SLC7A11 resulted in the induction of fatal oxidative stress through the impairment of glutathione synthesis and antioxidant mechanisms in HCC models, it exerted a substantial influence on the emergence of resistance to sorafenib in HCC cells (32). The oncogenic kinase AURKA is of significant importance in the control and coordination of the process of mitosis, as well as the proper segregation of chromosomes (33). In HCC, AURKA is frequently amplified and hyperactivated, leading to aberrant proliferation and survival signaling (34). Inactivation of AURKA has shown significant antitumor efficacy in HCC models, either as a monotherapy or in combination with sorafenib (35,36). NQO1 is a cytoprotective enzyme against oxidative stress and causes chemotherapy resistance through antioxidant mechanisms (37). In the context of liver cancer, previous study has documented a significant 18-fold increase in NQO1 expression in HCC compared to healthy liver tissue (38). The upregulation of NQO1 has been identified as a robust and autonomous biomarker for prognostic assessment in HCC, as well as a facilitator of enhanced inhibition of apoptosis in HCC cells through the SIRT6/AKT/XIAP signaling pathway (39). Elevated levels of NRF2/NQO1 in primary HCC are linked to the size of the tumor, elevated α-fetoprotein, and increased levels of des-γ-carboxy-prothrombin. Additionally, the excessive expression of NRF2/NQO1 is linked to numerous instances of intrahepatic recurrences and served as a separate risk factor for unfavorable prognosis. According to the research, focusing on the activity of NQO1 might be a viable approach (38).
In order to enhance our comprehension of the underlying biological functions and pathways associated with HCC, we conducted GO and GSEA analyses to explore the interactions among the 59 DRG-FRG genes. Using GO analysis, it was found that the 59 DRG-FRG genes are linked to various biological functions and signaling pathways. These have a significant impact on cancer development and progression, especially in HCC (40,41). Furthermore, GSEA revealed that the 59 DRG-FRG genes are predominantly associated with pathways closely linked to cancer and metabolism. One of the pathways significantly impacted by this signature is aminoacyl-tRNA biosynthesis, which plays a crucial role in protein synthesis and translation (42). This finding suggests that the DRG-FRG-based signature may influence the regulation of aminoacyl-tRNA formation, which is essential for the proper functioning of cellular processes. Additionally, the B cell receptor signaling pathway is identified as a key pathway influenced by the DRG-FRG-based signature, highlighting its involvement in controlling immune responses and promoting the activation and proliferation of B cells (43). This suggests that the signature may have a role in modulating immune-related functions, potentially affecting the immune response against cancerous cells. Another noteworthy pathway affected by the DRG-FRG-based signature is the cytosolic DNA-sensing pathway, which plays a pivotal role in innate immune responses by recognizing and responding to foreign DNA (44). The involvement of the signature in this pathway implies its potential role in regulating the cellular response to DNA damage, infection, and potentially cancer development. This underscores the importance of GO and GSEA analysis in identifying disease-relevant biological functions and signaling pathways, providing a foundation for researchers to explore the mechanisms of HCC formation and progression.
HCC is a prototypical tumor that arises due to inflammation, resulting in an immunosuppressive microenvironment. Consequently, the utilization of immunotherapy emerges as a promising approach. A comprehensive investigation conducted by Sangro et al. delved into the immunobiology of liver cancer, highlighting the promising prospects of immunotherapeutic interventions (45). In their study, Donisi and colleagues explore the significance of immune checkpoint inhibitors in the treatment of HCC, they also highlight the potential of synergistic combinations involving immune checkpoint blockade and tyrosine kinase inhibitors, anti-angiogenics, or other immunotherapies as promising and emerging therapeutic approaches for HCC (46).
Limitations of this study include lack of independent validation and functional investigation of the identified genes and pathways. The prognostic model requires further confirmation in large HCC cohorts. Additionally, the detailed molecular mechanisms connecting these biomarkers with HCC biology need to be determined. Despite these limitations, the study has significant implications for the understanding and treatment of HCC. The identification of the 11 genes and the construction of the risk score model provide valuable tools for the prognosis and stratification of HCC patients. These tools could potentially be used to guide the selection of therapeutic strategies and to monitor disease progression. Moreover, the identified genes provide potential targets for therapeutic intervention in HCC. The development of drugs targeting these genes could potentially improve the efficacy of HCC treatment and overcome drug resistance, which is a major challenge in the treatment of HCC.
Conclusions
In summary, this study offers significant contributions to the understanding of the molecular mechanisms involved in HCC and identifies promising targets for therapeutic intervention. The results emphasize the significance of integrating bioinformatics analysis with experimental and clinical research endeavors to enhance the prognosis and treatment of HCC. To confirm these results and explore their potential clinical applications, further experimental studies are required.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1594/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1594/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1594/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Ganesan P, Kulik LM. Hepatocellular Carcinoma: New Developments. Clin Liver Dis 2023;27:85-102. [Crossref] [PubMed]
- Liu X, Nie L, Zhang Y, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol 2023;25:404-14. [Crossref] [PubMed]
- Zheng P, Zhou C, Ding Y, et al. Disulfidptosis: a new target for metabolic cancer therapy. J Exp Clin Cancer Res 2023;42:103. [Crossref] [PubMed]
- Zhang R, Kang R, Tang D. Reductive cell death: the other side of the coin. Cancer Gene Ther 2023;30:929-31. [Crossref] [PubMed]
- Zhao S, Wang L, Ding W, et al. Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework. Front Endocrinol (Lausanne) 2023;14:1180404. [Crossref] [PubMed]
- Machesky LM. Deadly actin collapse by disulfidptosis. Nat Cell Biol 2023;25:375-6. [Crossref] [PubMed]
- Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021;12:599-620. [Crossref] [PubMed]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266-82. [Crossref] [PubMed]
- Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019;12:34. [Crossref] [PubMed]
- Wu J, Wang Y, Jiang R, et al. Ferroptosis in liver disease: new insights into disease mechanisms. Cell Death Discov 2021;7:276. [Crossref] [PubMed]
- Bekric D, Ocker M, Mayr C, et al. Ferroptosis in Hepatocellular Carcinoma: Mechanisms, Drug Targets and Approaches to Clinical Translation. Cancers (Basel) 2022;14:1826. [Crossref] [PubMed]
- Hu FF, Liu CJ, Liu LL, et al. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Brief Bioinform 2021;22:bbaa176. [Crossref] [PubMed]
- Zhu K, Liu X, Deng W, et al. Identification of a chromatin regulator signature and potential candidate drugs for bladder cancer. Hereditas 2022;159:13. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Edwards DN, Ngwa VM, Raybuck AL, et al. Selective glutamine metabolism inhibition in tumor cells improves antitumor T lymphocyte activity in triple-negative breast cancer. J Clin Invest 2021;131:e140100. [Crossref] [PubMed]
- Zheng L, Yang Q, Li C, et al. Ubiquitin-Specific Peptidase 8 Modulates Cell Proliferation and Induces Cell Cycle Arrest and Apoptosis in Breast Cancer by Stabilizing Estrogen Receptor Alpha. J Oncol 2023;2023:8483325. [Crossref] [PubMed]
- Wang S, Song Z, Tan B, et al. Identification and Validation of Hub Genes Associated With Hepatocellular Carcinoma Via Integrated Bioinformatics Analysis. Front Oncol 2021;11:614531. [Crossref] [PubMed]
- Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226-1239.e4. [Crossref] [PubMed]
- Ye W, Shi Z, Zhou Y, et al. Autophagy-Related Signatures as Prognostic Indicators for Hepatocellular Carcinoma. Front Oncol 2022;12:654449. [Crossref] [PubMed]
- Wang Z, Zhu J, Liu Y, et al. Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma. J Transl Med 2020;18:67. [Crossref] [PubMed]
- Chang YS, Lee YT, Yen JC, et al. Long Noncoding RNA NTT Context-Dependently Regulates MYB by Interacting With Activated Complex in Hepatocellular Carcinoma Cells. Front Oncol 2021;11:592045. [Crossref] [PubMed]
- Zhou X, Huang JM, Li TM, et al. Clinical Significance and Potential Mechanisms of ATP Binding Cassette Subfamily C Genes in Hepatocellular Carcinoma. Front Genet 2022;13:805961. [Crossref] [PubMed]
- Wang G, Guan J, Yang Q, et al. Development of a Bile Acid-Related Gene Signature for Predicting Survival in Patients with Hepatocellular Carcinoma. Comput Math Methods Med 2022;2022:9076175. [Crossref] [PubMed]
- Cihalova D, Staud F, Ceckova M. Interactions of cyclin-dependent kinase inhibitors AT-7519, flavopiridol and SNS-032 with ABCB1, ABCG2 and ABCC1 transporters and their potential to overcome multidrug resistance in vitro. Cancer Chemother Pharmacol 2015;76:105-16. [Crossref] [PubMed]
- Buschauer S, Koch A, Wiggermann P, et al. Hepatocellular carcinoma cells surviving doxorubicin treatment exhibit increased migratory potential and resistance to doxorubicin re-treatment in vitro. Oncol Lett 2018;15:4635-40. [Crossref] [PubMed]
- Hamed AR, Yahya SMM, Nabih HK. Anti-drug resistance, anti-inflammation, and anti-proliferation activities mediated by melatonin in doxorubicin-resistant hepatocellular carcinoma: in vitro investigations. Naunyn Schmiedebergs Arch Pharmacol 2023;396:1117-28. [Crossref] [PubMed]
- Yang L, Zhang W, Yan Y. Identification and characterization of a novel molecular classification based on disulfidptosis-related genes to predict prognosis and immunotherapy efficacy in hepatocellular carcinoma. Aging (Albany NY) 2023;15:6135-51. [Crossref] [PubMed]
- Xu J, Wu X, Wang X. Ferroptosis-Related Genes with Regard to CTLA-4 and Immune Infiltration in Hepatocellular Carcinoma. Biochem Genet 2023;61:687-703. [Crossref] [PubMed]
- Chen Q, Zheng W, Guan J, et al. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ 2023;30:137-51. [Crossref] [PubMed]
- Li Y, Yang W, Zheng Y, et al. Targeting fatty acid synthase modulates sensitivity of hepatocellular carcinoma to sorafenib via ferroptosis. J Exp Clin Cancer Res 2023;42:6. [Crossref] [PubMed]
- Du R, Huang C, Liu K, et al. Targeting AURKA in Cancer: molecular mechanisms and opportunities for Cancer therapy. Mol Cancer 2021;20:15. [Crossref] [PubMed]
- Li G, Tian Y, Gao Z. The role of AURKA/miR-199b-3p in hepatocellular carcinoma cells. J Clin Lab Anal 2022;36:e24758. [Crossref] [PubMed]
- Zheng D, Li J, Yan H, et al. Emerging roles of Aurora-A kinase in cancer therapy resistance. Acta Pharm Sin B 2023;13:2826-43. [Crossref] [PubMed]
- Su WL, Chuang SC, Wang YC, et al. Expression of FOXM1 and Aurora-A predicts prognosis and sorafenib efficacy in patients with hepatocellular carcinoma. Cancer Biomark 2020;28:341-50. [Crossref] [PubMed]
- Zhang K, Chen D, Ma K, et al. NAD(P)H:Quinone Oxidoreductase 1 (NQO1) as a Therapeutic and Diagnostic Target in Cancer. J Med Chem 2018;61:6983-7003. [Crossref] [PubMed]
- Shimokawa M, Yoshizumi T, Itoh S, et al. Modulation of Nqo1 activity intercepts anoikis resistance and reduces metastatic potential of hepatocellular carcinoma. Cancer Sci 2020;111:1228-40. [Crossref] [PubMed]
- Zhao W, Jiang L, Fang T, et al. β-Lapachone Selectively Kills Hepatocellular Carcinoma Cells by Targeting NQO1 to Induce Extensive DNA Damage and PARP1 Hyperactivation. Front Oncol 2021;11:747282. [Crossref] [PubMed]
- Vaghari-Tabari M, Ferns GA, Qujeq D, et al. Signaling, metabolism, and cancer: An important relationship for therapeutic intervention. J Cell Physiol 2021;236:5512-32. [Crossref] [PubMed]
- Garcia-Lezana T, Lopez-Canovas JL, Villanueva A. Signaling pathways in hepatocellular carcinoma. Adv Cancer Res 2021;149:63-101. [Crossref] [PubMed]
- Rubio Gomez MA, Ibba M. Aminoacyl-tRNA synthetases. RNA 2020;26:910-36. [Crossref] [PubMed]
- Tanaka S, Baba Y. B Cell Receptor Signaling. Adv Exp Med Biol 2020;1254:23-36. [Crossref] [PubMed]
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016;17:1142-9. [Crossref] [PubMed]
- Sangro B, Sarobe P, Hervás-Stubbs S, et al. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:525-43. [Crossref] [PubMed]
- Donisi C, Puzzoni M, Ziranu P, et al. Immune Checkpoint Inhibitors in the Treatment of HCC. Front Oncol 2021;10:601240. [Crossref] [PubMed]


