
CRISPR/Cas9 engineering offers new opportunities to model pancreatic ductal adenocarcinoma development
Pancreatic ductal adenocarcinoma (PDAC) is a dismal disease. The 5-year survival is below 5%, which is due to a combination of aggressive disease progression, late diagnosis and limited treatment options (1). Consequently, PDAC is predicted to be the second-largest contributor to cancer related deaths by 2020 (2). New approaches and models to accelerate therapeutic development and improve patient outcome are therefore urgently needed.
The common genetic aberrations of PDAC have been identified, where activating mutations in KRAS in combination with loss of function in the tumor suppressors SMAD4, CDKN2A and TP53 represent the most frequently occurring aberrations (3-6). Moreover, genomic rearrangements are typically observed in PDAC, where recent studies have grouped tumors dependent on the extent of rearrangements (4). Further, expression analysis in both primary cell lines as well as in tumors have identified a number of sub-classes with differing prognosis (7-9). Although there are still no targeted therapies clinically available for these aberrations, new opportunities may arise when less frequent mutations are categorized by their molecular function (6,10).
Our current understanding of the role whereby mutant KRAS drives PDAC development and how loss of function in SMAD4, CDKN2A and TP53 augment malignant progression stems from diligent analysis of preclinical models, where genetic engineered mouse models (GEMMs) have played an instrumental role (11,12). However, while several aspects of human PDAC are well recapitulated by GEMMs, including stromal desmoplasia, metastasis and genomic instability, other aspects are not recapitulated similarly. For example, in most GEMMs genetic inserts are driven by tissue specific promoters that induce expression in most cells during pancreas development. This results in multi-focal disease with short latency, which is in contrast to the human disease etiology that exhibits a stochastic development of disease with longer latency. Moreover, manipulation of multiple targets is cumbersome and requires large cohorts to derive sufficient high numbers of animals for studies. Consequently, animal studies of interplay between multiple co-occurring genetic aberrations have been limited.
To address these issues, Maresch and colleagues have developed an elegant method to introduce specific genetic aberrations in the adult pancreas in vivo using the CRISPR/Cas9 system for genome engineering (Figure 1) (13).
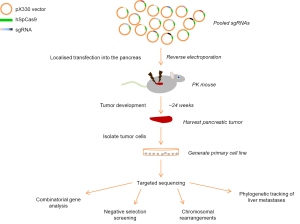
Building on previous experience with in vivo delivery of plasmid DNA, the authors demonstrate that injection of DNA into the pancreas of adult mice, followed by electroporation, initially targets an average of 750 cells. While some cells undergo apoptosis and are removed by a local inflammatory response, an average of 120 cells survive long-term, demonstrated using the Cre reporter model Rosa26mT/mG. As such, electroporation of plasmid DNA only targets a small fraction of pancreatic cells, which may be a better approximation of the stochastic nature of the human disease.
To determine the effect of a number of tumor suppressor genes that display different frequency of loss of function in human PDAC, the authors generated individual guide RNA plasmids to target 13 tumor suppressor genes and 2 for the ‘neutral’ Rosa26 locus. When injected into the pancreas of animals bearing pancreas-specific expression of the oncogenic driver KrasG12D (PK: Ptf1aCre/+; KrasLSL-G12D/+), the resulting animals developed tumors at accelerated rate with different histopathologic characteristics as well as overt liver metastasis. Sequencing of the targeted alleles from isolated cell lines clearly demonstrated high frequency indels at multiple CRISPR/Cas9 sites where 7 to 14 of the 15 targets showed simultaneous mutations. Interestingly, the authors didn’t identify loss of function in Brca2, suggesting that the methodology may be used for negative selection screening.
Due to the relatively low frequency of cells targeted by electroporation, the authors hypothesized that the model also could be used for phylogenetic tracking of metastatic disease. Comparing the targeted allele frequency of the guide RNAs in tumor cell clones from 8 regions of the primary tumor and matching liver metastasis, the authors noted that while there was only a minor 5% contribution of clone 1 (of 2) in the primary tumor, the relative contribution was 50% between the clones at the metastatic sites.
Chromosomal rearrangements are a common feature of PDAC, where both intra-chromosomal and inter-chromosomal deletions, unbalanced translocations and chromothripsis are observed. Genome engineering by CRISPR/Cas9 has previously been shown to result in intra- and inter-chromosomal deletions and therefore an extensive analysis of these aberrations were undertaken across tumors and isolated cell lines. Inter-chromosomal deletions were frequently observed, with 3 out of 6 tumors displaying large deletions. Moreover it is noteworthy that these deletions frequently target tumor suppressors. Inter-chromosomal translocations were less common with only one event/tumor. Chromosomal aberrations have previously been observed in GEMMs of PDAC (11), albeit the nature and molecular understanding of these are still underexplored. Taken together these data suggests this model recapitulates critical aspects of genome instability that may promote disease progression.
Overall the new model offers important complementarity to already available pre-clinical models and there are a plethora of unaddressed questions that can be now addressed. For example, it would be interesting to address whether tumors of different histopathology characteristics observed in the model display significant differences in their allele frequency of the targets and/or whether additional mutations have accumulated. It would also be highly informative to determine how well the overall mutational and neo-antigen burden in this model compares to other GEMMs as well as human PDAC. A subject not touched upon in this publication is the stromal reaction, where PDAC is characterized by a highly desmoplastic reaction. Whether some aspects of the human stromal reaction are better represented by this model remains to be described. Finally, there is now an unique opportunity to rapidly compare mutational spectrum with therapeutic response, a much-needed element to evaluate novel regimes for clinical translation.
Acknowledgments
Funding: This work was funded by a Cancer Research UK Career Establishment Award (C37293/A12905) and a Cancer Research UK Institute Award (A19258).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Gang Wang, PhD (Department of Pancreatic and Biliary Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.08.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hidalgo M. Pancreatic cancer. N Engl J Med 2010;362:1605-17. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6. [Crossref] [PubMed]
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Chang DK, Grimmond SM, Biankin AV. Pancreatic cancer genomics. Curr Opin Genet Dev 2014;24:74-81. [Crossref] [PubMed]
- Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7:469-83. [Crossref] [PubMed]
- Tuveson DA, Shaw AT, Willis NA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 2004;5:375-87. [Crossref] [PubMed]
- Maresch R, Mueller S, Veltkamp C, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun 2016;7:10770. [Crossref] [PubMed]


