
Utilizing signature-score to identify oncogenic pathways of cholangiocarcinoma
Introduction
Cellular functions and biological processes are determined by cell-type-specific transcriptional programs. Thus, knowledge of the transcriptional derangements present in tumors can be used to predict altered function and identify pathways and processes that drive tumor growth. These altered functions and pathways can be targeted and exploited for therapeutic intervention. The inherent heterogeneity and complexity of tumors is reflected by a significant degree of variability between gene signature sets (GSSs) obtained from similar tumor types. Gaining a broad-based view of pathways and processes associated with specific tumor type, thus requires further analysis such as group comparison or clustering of differentially expressed genes.
Different scoring methods have been developed to quantify the activities of GSSs associated with different disease states. Hiromichi and his colleagues used averaged Z-value of genes as a “deregulation index” to score GSS (1). Barbie et al. utilized Kolmogorov-Smirnov (KS) statistics to extend the Gene Set Enrichment Analysis (GSEA) (2) to project GSSs’ expression to score single sample (3). Pearson correlation of comparing the directions of expression fold changes of member genes was also proposed for GSS scoring (4). Among these methods, Z-value and KS statistics are effective only when member genes have consistent directional changes in expression. However, these underlying assumptions do not necessarily hold true for the following reasons; (I) GSSs derived from genome-wide profiling experiments contain both up- and/or down-regulated components, which differ according to their cells and tissue of origin, and (II) the magnitude of gene expression alterations reflect magnitude of the response under any given experimental condition or disease state (e.g., benign vs. invasive tumors). In addition, many of these methods only consider the expression pattern (change in direction; up or down) and regard every gene with equal weight in the scoring scheme. In other words, the significance of each gene in the signature set is neglected. Recognizing these issues, we developed a unique scoring method to reconcile all of these factors within a GSS to generate a quantitative score.
To demonstrate the effectiveness of the method, we applied S-score to cholangiocarcinoma (CAC), the second most common primary liver cancer after hepatocellular carcinoma (HCC) (5). We applied our analysis to CAC as it is a clinically silent malignancy and as such, has an exceptionally dismal prognosis (5). Patients show a median survival rate of ~12 months with a 5-year survival rate of only 5-10% due to the presence of locally invasive or advanced metastatic disease at the time of diagnosis, precluding the possibility of resection. In cases where resection is possible, the high mortality rate reflects a high rate of post-surgical relapse and poor response to chemotherapy (6-8). While genetic alterations in KRAS, TP53, BRAF and EGFR have been reported for CAC (9-11), there has been a relative lack of systematic exploration. Thus, other alteration of important oncogenes and tumor suppressor genes were largely remain unclear.
To characterize individual CACs and identify oncogenic pathways and processes that drive biliary transformation, we used S-score, a novel signature scoring method adapted from our previously used scoring system (12) to gene expression profiles of CACs. S-score is different from previously used methods in that it concurrently evaluates both up- and down-regulated components of a GSS through a sign function and adjusts member genes’ weights by differential expression significance, or P-value. As proof of principle and to demonstrate the accuracy and robustness of S-score, we conducted a thorough and comparative simulation under various conditions. In addition, the qualitative boundary of the S-score was established to provide a measure of the dynamic range of a GSS’s activity (or the status of a oncogenic pathway). Finally, we applied S-score to two expression data sets of CAC containing 31 GSSs to identify putative oncogenic alterations, pathways and processes. Using S-score, we identified several putative oncogenic alterations associated with CAC as well as differentially expressed pathways associated with CAC and HCC.
Merterials and methods
Signature score (S-score)
In order to project gene expression levels of a set of genes (a GSS) to a scalar score, a scoring method, adapted from our previous work (12), was developed to quantify each sample. We briefly describe the method as follows.
Suppose there are N genes in a GSS derived from a case-control gene expression profiling experiment. The case-control experiment contains M expression arrays, and represents the gene expression level of the jth gene from ith sample. Assuming there are W testing samples, and let , where is the log2-transformed expression level of gene j in the testing sample . To assess the activity of a gene set, we apply following operator,
Where is the z-score of a test sample, or
Where and are mean and standard deviation of gene j across all M case-control experiment samples. Similar approach has been utilized in Eq. [1] and Eq. [4] by replacing z-score with a Pearson correlation coefficient to case-control status in Eq. [1].
To incorporate the effect of differential expression significance and directionality, let ρj be the fold change of gene j between case and control, pj be the P-value of Student t-test of gene j in the case-control experiment from which the GSS was derived. As defined before, N is the number of genes in the GSS. The Signature Score (S-score) of the testing sample l is defined as,
where
and
Eq. [4] is set that if p>0.1, or , then =0, and K Eq. [5] is the sum of all weight in Eq. [3]. p* is designed to remove genes that has no test significance and weight more to differentially expressed genes, but limited to no more than 3× weight when p is less than 10-4. Note in Eq. [3], if a test sample was profiled with the same microarray platform as that in case-control experiment, then,
Where and are the standard deviations of case and control, respectively, and and are the numbers of cases and controls in the experiment, respectively. However, if the test samples were profiled with a different microarray platform, and are mean and standard deviation of gene j across all W arrays of test samples, as an estimate of training samples.
Notice that S-score , defined by Eq. [3], is positive when its standardized expression value z and the fold change of the corresponding gene in case/control samples have the same directional sign (positive or negative).
Qualitative boundary of S-score and no-call region
In concept, samples with positive S-scores that have expression patterns similar to case experiments can be classified as having an “active status” while those with negative scores can be classified as having an “inactive status”. However, it is necessary to define a reliable prediction interval of active/inactive status in the whole dynamic range of S-score. In this study, we define the boundary of active/inactive statuses to be the 99% prediction interval under the null hypothesis that all signature genes are randomly distributed and not correlated to case/control in the conditional experiment. Let zj,i be normally distributed with N[0, 1], and there are n genes and total of M microarrays balanced in active and inactive groups in the conditional experiment. The prediction interval is defined as,
where we assumed that the S-score of random gene expression is approximately normally distributed [with standard cumulative distribution function Φ(•)] governed by the Large Number Theorem, μs and σs are mean and standard deviation of which can be determined by simulation. For z* to be N[0, 1] in Eqs. [2-3], it can be easily shown that . The null hypothesis will be rejected and the status could be identified if the S-score exceeds or is below the L0.01, or samples with an S-score between L0.01 will not be classified in neither active nor inactive status (no-call group) to control the call-error below 1% due to random events. In Supplementary Materials section S3, we also provided a range of L values under various sample sizes, and a specific example for the determination of oncogene status with statistical confidential through our model Eq. [7]. Also, if the “no-call” region defined by L0.01 covers entire dynamic region of a given GSS, the GSS will be classified as not involved in the biological system under study.
Generation of gene signature sets
A total of 31 GSSs were generated from data obtained from sources listed in Table S1 of the Supplementary Materials, The number of genes, data sources, and related studies were listed in Table S1 in the Supplementary Merterials. The cell cycle signature (CCS) that was used to define tumor cell proliferation was generated from a cell-cycle signature master set derived from genes preferentially expressed in actively cycling cells as well as genes that were repressed following growth inhibition (13,14). The wound healing signature (WHS) was derived from the gene sets response to serum stimulations (15). The KRT19 and EpCAM GSSs were generated from differentially expressed genes previously identified in human or rat HCCs (16,17). c-Met and TGFβ signatures were generated from the expression profiles of wild-type and gene knockout mouse hepotocytes (18,19). The Shh (Sonic Hedgehog) signature was derived from an Shh-subtype of medulloblastoma (20). The RAF, MEK, Erbb2, and EGFR+EGF signatures were derived from genes found to be overexpressed in MCF-7 cells (21). The signatures of Src, E2F1, STAT3, p53, p63, Myc, ER, AKT, PI3K, Her2, TNF, IFNα, IFNβ, β-Catenin, EGFR, TGFβ, PR, ER, and Ras were derived from the study by Bild et al. (22).
Data set
Three datasets were used in this paper. The first data set which contains 94 embryonal rhabdomyosarcomas (eRMS) and 18 normal skeletal muscles, was download from caArray (https://array.nci.nih.gov/caarray/project/details.action?project.experiment. publicIdentifier=trich-00099) that we used in (12). Data set GSE26566 and GSE15765 were downloaded from GEO database. GSE26566 contains 104 cholangiocarcinoma specimens (CAC), 59 normal liver tissues (NL), and 6 normal intrahepatic bile ducts (IBD). GSE15765 contains 70 hepatocellular carcinoma (HCC), 13 cholangiocarcinoma (CAC), and 7 samples of mixed type of combined HCC and CAC (CHC).
Results
Performance and robustness of S-score
To demonstrate the performance of S-score, the eRMS data, discussed in (12), were used to compare the capability of S-score against the other three scoring methods, ES-score (3), averaged Z-score (1), and Pearson correlation (4). The result showed that (see Figure S1), among 4 methods, S-score and correlation-based method have the capacity to integrate up- and down-regulated gene sets. The two methods can accurately project expression pattern to a signature score, with the suppression of the noisy effect of unrepresentative genes in the GSS, if they exist (Details in Supplementary Materials section S1).
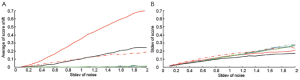
To further estimate the robustness of the scoring methods, we performed simulations where we added random noises into one positive sample as marked in Figure S1A. The simulation was performed on one of GSSs, p53off, derived by comparing cancer samples with p53 loss of function vs. samples with normal p53 function. For Z-score method, we separate p53off GSS into two sets: p53off-up and p53off-down where up- and down-regulated genes were grouped, respectively. The mean of added noises was set to zero and the strengths (standard deviation) were increased from 0.1 to 2 times the standard deviation of each gene. Each condition was simulated 1,000 times and then calculations were made for the mean shift and standard deviation of each score. The result, shown in Figure 1, revealed that S-score and Z-score have the smallest score shift (all close to zero), regardless of the standard deviation of added noise. The mean shift of correlation is similar to ES-score in p53off-up and p53off-down that increase with the standard deviation of noise. Among these methods, ES-score with p53off-up gene set has the largest mean shift in the simulations, indicating worst robustness under noisy conditions (Figure 1A). The standard deviations of all four test scores were varied at a similar range (Figure 1B), with those of ES-score with p53off-up genes and correlation slightly lower than other methods. However, these two methods also have the largest mean-shift under noisy conditions.
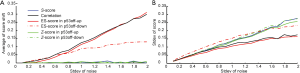
The robustness of S-score was further demonstrated in Supplementary Materials Section S2 when a negative sample was also applied for the simulation. Similarly, S-score have a small mean shift and a similar range of standard deviation, as shown in Figure S2. Combining these two observations from simulations, S-score and Z-score have the best robustness (mean shift close to zero) against noises in the test samples, while S-score has the ability of suppressing non-performing genes in a GSS.
Identification of oncogenic pathways of cholangiocarcinoma
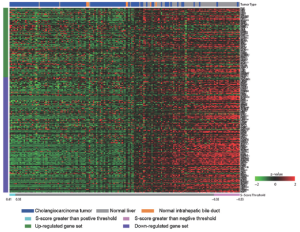
Cholangiocarcinoma (CAC) is one of malignent cancers in the world, however, only mutations of gene Kras, p53 and Raf have been reproted. Other driving oncogentic pathways are still unclear. Here we ultized S-score method to investgate the activities of oncogenic pathways of CAC through gene expression profiles. To identify the driving oncogene, oncogenic signatures of known oncogenes and the actived pathway have been reported in the HCC, were collected. Total 31 oncogenic signatures was applied to a gene expression data set of CAC, GSE26566, based on the S-score to estimate the activities of the pathways. The data set contains 104 CAC specimens, 59 normal liver tissues (NL), and 6 normal intrahepatic bile ducts (IBD). Three gene signature sets (GSSs), p63, AKT, and PR, were excluded due to none or only one sample passed the qualitative boundary L0.01. The c-Met signature was also excluded because most of the up or down-regulated genes in the GSS were highly expressed in NL samples that lost the trend of expression direction in the original case-control study (Figure S3 in Supplementary Materials). S-scores for each sample and each GSS were generated and then a Student t-test was used to identify the statistical significance of differential oncogenic pathways. Using P-values of t-test smaller than 0.05 as the criteria, 13 of 27 signatures were shown the differential activities between CAC and the two normal tissues (Figure 2 and S4 in the Supplementary Materials). Twelve out of 13 GSSs showed higher values of S-score in CAC. Only Src signature had lower value of S-score in CAC. On average, 21 CAC samples (20%) have identified as “on” status of signatures, which labeled as cyan in the Figure 2. KRT19 have the highest identified percentage: 73 of 104 CAC samples were identified as “on” status of KRT19 with an S-score greater than L0.01 (0.44). The result was consistent with previous reports that KRT19 is highly expressed in CAC (23-25).
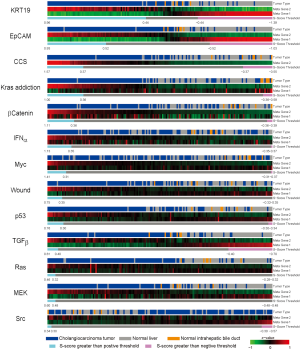
To investigate the co-regulation of the oncogenic signatures and the subtypes of CAC, a two-way hierarchical clustering was performed. The result identified two major clusters of signatures’ co-activity. The signature cluster A, which labeled with blue color in Figure 3A, manifested the correlation of β-Catenin, Ras, CCS and wound signatures. The cluster B which labeled with red color exhibited the co-expression of TGFβ, KRT19, and EpCAM. The values of Pearson correlation between two GSSs were further substantiated the clustering results (Figure 3B). The data also showed that IFNα and Src signatures were active in an independent manner without correlation with other signatures. On the other hand, 6 clusters of CAC samples were identified by using hierarchical clustering (Figure 3). The cluster 1 has the strongest activities (except of IFNα and Src signatures) among all the clusters; Cluster 2 has less activity of signature cluster A; Cluster 3 has less expression of p53 and MEK; Cluster 4 has high activities of signature cluster B with different combination of other signature activities; Cluster 5 has moderate activities of signature cluster B but had low activities in most of other signatures; and Cluster 6 shows low activities of all signatures (except INFα and Src). Clearly, our result demonstrated the heterogenetic characteristic of oncogenic background in CAC patients.

The oncogene associated gene ontology items of cholangiocarcinoma
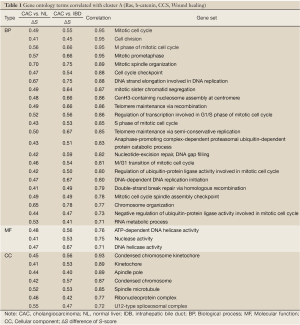
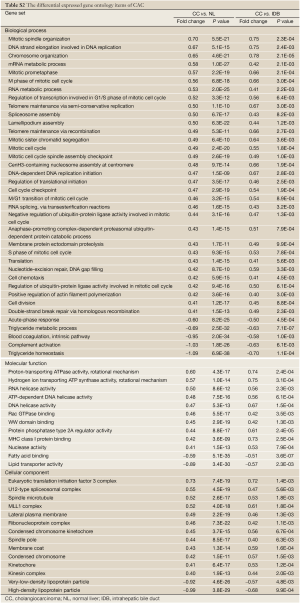
To understand the dysregulated function of CACs, we also investigated GSSs with Gene Ontology (GO) terms. Using log2-fold change >0.4 and P-value <0.01 in both CAC vs. NL and CAC vs. IDB comparisons as the selecting criteria, a total of 68 GO terms, containing 39, 12, and 17 terms in biological process (BP), molecular function (MF), and cellular components (CC), respectively, were shown significant differentially activities by S-score in CAC (Table S2 in Supplementary Materials). In the CAC associated GO items, 54 of 68 CAC associated GO items were up-regulated (32, 10, and 12 in BP, MF, and CC, respectively) and 14 were down-regulated (5, 2, and 2 in BP, MF, and CC, respectively). Several mitotic correlated terms, such as mitotic spindle organization, mitotic prometaphase, and M phase of mitotic cell cycle, were up-regulated in CACs. Also, RNA transcription related terms and telomere maintenance terms were up-regulated as well. Those results indicated CACs have strong cell cycle and RNA transcription activities comparing with normal NL or IDB tissues. We also observed down-regulation of GO terms of triglyceride homeostasis, complement activation, acute-phase response, and lipid transporter, indicating the potential loss of these functions in CAC cells.
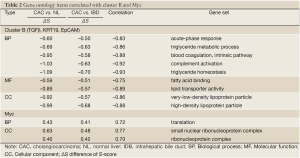
To investigate the relationship between the CACs associated oncogene pathways and functions, Pearson correlation was applied to the S-score values of the 12 oncogenes and 68 GO terms. A total 45 terms met the criteria of correlation coefficients >0.7. The cell cycle and mitotic related terms were highly correlated with the cluster A (Table 1). Also other terms of cell functions, such as mRNA metabolic, translation, and telomere maintenance via recombination, possessed positive correlation with signature cluster A. This association suggested the activities of β-Catenin and Ras, the oncogenes of the Cluster A, were related to activities of cell dividing, transcription, and translation in CACs. Similarly, due to the negative correlation of the Cluster B to two immune related terms, acute-phase response and complement activation, and lipoprotein related terms, such as lipid transporter activity and triglyceride homeostasis, suggested involvement of Cluster B in the suppression of the immune response and lipid transporter functions (Table 2).
Full table
Full table
Comparsion of cholangiocarcinoma and hepatocellular carcinoma
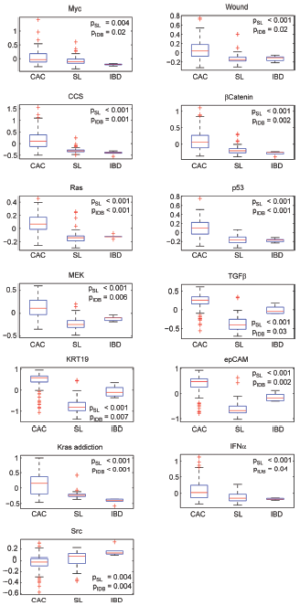
The hepatocellular carcinoma (HCC) with cholangiocarcinoma signature has been reported with worse prognosis (26). However, the differences between these two cancers in the oncogenic pathway are still unknown. A data set GSE15765 that contains 70 hepatocellular carcinoma (HCC), 13 cholangiocarcinoma (CAC), and 7 samples of mixed type of combined HCC and CAC (CHC) were used to investigate the differences with our oncogenic GSSs. By performing t-test between HCC and CAC samples, total 12 oncogenic GSSs were different with statistical significance P<0.05. Nine of 12 signatures have higher activities in CAC (Figure 4A) and 3 oncogenic GSSs, AKT, PI3K, and E2F1, showed greater levels in HCC (Figure 4B). Among the 9 highly expressed GSSs in CAC, 6 were overlapped with the analysis of GSE26566: MEK, Ras, k-ras addiction, KRT19, EpCAM, and TGFβ. The GSSs of Her and Erbb2, which were signature of the same gene, ERBB2, but derived from different experiments, were both shown highly expressed in CAC.
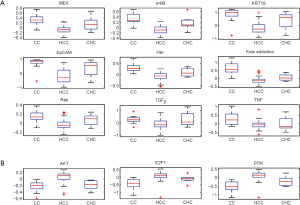
In addition to HCC, the mixed tumor type CHC showed mixed oncogenic GSSs S-score range (activities) in all 12 GSSs, as we expected. To summarize our observation in Figure 4, HCC and CAC have distinct activities of oncogenic pathways, and they can be efficiently utilized for tumor classification, instead of individual genes. However, the topic is beyond the scope of this paper.
Discussion
We proposed a novel tumor scoring system, S-score, to quantify oncogenic GSSs in tumor comparison. By considering the directions of fold change and P-values of GSSs, S-score provides an effective scoring method for each tumor with accuracy and robustness, as demonstrated in the Result Section. Not only for quantitation, by estimating the confidential boundary L0.01, the qualitative status of GSSs of each sample can also be determined. The boundary L0.01 was also utilized to filter out the signatures of which distribution of S-scores are relatively small. For example, the results of the signatures p63, AKT, and PR, were excluded in the analysis of GSE26566 after filtering by L0.01 to avoid the situation of small variance of S-score, causing instable t-test significance.
Through S-score, we expect further insight into gene signature set analysis of a tumor to understand the regulation and function of pathway of cancer biology. Using expression profiles of CACs, 12 oncogenic GSSs were shown with high activities in CAC comparing with two normal tissues (Figure 2), consistent with the observations reported earlier in literatures. Kras and p53 have been reported with high frequency of mutation in the CAC patients (9,10). Mutation of SMAD4, which was a downstream gene of TGFβ pathway, also has been identified in a proportion of CAC patients (27). Immunohistochemical analysis of β-Catenin in a previous study showed positive staining in cytoplasm and/or nucleus in 58.3% of 24 CAC samples (28). KRT19, EpCAM were highly expressed in biliary epithelial cells, and have been report as the markers of hepatic progenitor cells with β-Catenin (16,17). Our data indicated that KRT19 and EpCAM were coexpressed with TGFβ in CAC. Although no mutations or alteration of expression of MEK in CAC have been reported earlier, but a phase II clinical trial have been reported with 12% response rate of selumetinib, a MEK inhibitor, for CAC patients (29).
Our analysis also showed that cell-cycle signature (CCS) and wound healing signature were highly correlated with GSSs of Myc and β-Catenin in CACs (Figure 3). The high scores of CCS and wound signatures indicated high proliferation and invasion in most of CAC samples, perhaps Myc and/or β-Catenin play important roles in the tumor progression of CAC.
Two Ras related GSSs were showed associated with CACs (Figure 2). However, the correlation of the GSSs of Ras and K-ras addiction was low (Figure 3). The Ras GSS was derived from gene affected by Hras overexpression. The Kras addiction was derived from the K-ras dependent cell lines, which mean K-ras played a dominant role of the proliferation signal in the cell lines. While we don’t expect two GSSs have the similar S-scores for all CACs, we assume other oncogenes, perhaps not Kras, may play the more significant roles even in the Kras mutated CAC tumors. By analyzing all oncogenic GSSs, our analysis may provide additional clue to other oncogenes that CACs may addict to.
The cluster analysis in the study identified 6 subclasses of CAC patients with different combinations of oncogene activities. This information can provide a useful molecular evidence for predicting prognosis and making treatment decisions, especially for target drugs. For example, the patients with higher MEK activity could have high probability to response to MEK inhibitor (29). Additional treatment strategies can be designed to target other oncogenic pathways with the potential to increase the drug response rate toward the goal of personalized medicine. For example, in the CACs cluster 6, the low activities of GSSs not only observed in cluster B (TGFβ, KRT19, and EpCAM) but also in all other GSSs except Src. We assumed that there were other oncogenic pathways (GSSs) not collected in the study. As we can see from Figure 3A, more GSSs are needed to uncover the known oncogenes in patients of clusters 2-4.
The correlation between GSSs and GO terms were also investigated in this study. The cell cycle related GO terms were associated with GSSs of the Cluster A. The result further confirmed that two oncogenes, Myc and β-Catenin, might play an important role in the cell cycling of CAC cancer cells. In addition, the two oncogenes were also associated with the activities of other essential function of cells, such as mRNA metabolic, translation, and telomere maintenance via recombination, suggesting the causal relationships might exist between the two oncogenes and the functional consequences. Our analysis also indicated the activities of the Cluster B (EpCAM, KRT19, and TGFβ) was negatively correlated to two immune functions, acute-phase response and complement activation. Since the chronic inflammation has been identified as an important factor of carcinogenesis (30,31), not only these three genes are used as markers for diagnosis of CACs, but also are involved in the carcinogenesis of CAC due to their ability to suppress immune functions.
Comparing to HCC at oncogenic GSS level, CACs have higher activities of MEK and Her2 while HCCs show high activities of AKT, PI3K and E2F1. Timothy H. et al. reported that PI3K/AKT signaling but not MEK blocks E2F1-induced apoptosis and switch E2F1 to proliferative function (32). Aberrant Her2 expression in the CAC patients with poor prognosis has been described by Jesper A. et al. (33). Our results, combined with literature reports, clearly suggested the proliferation signaling between HCC and CAC is different. Moreover, HCC patients with high activities of EpCAM, KRT19, and TGFβ have been reported with poor prognosis. These findings supported the clinical observations that CAC is malignant with relatively short survival rate.
The progression of cancer can be reasoned from different kinds of alteration in the genome, such like mutations, copy number variations, and epigenetic modifications. The consequences of these alterations will induce changes of gene expression and then to the protein expression level in their pathways. Utilizing our S-score method that estimates the expression pattern of the oncogenic GSSs derived from in vivo or in vitro experiments, the putative oncogenic pathways could be uncovered as we demonstrated in the two CAC data sets. Some of the oncogenic pathways we identified were well known and some are yet characterized in CAC. Those data demonstrated that S-score is a rational strategy to explore novel oncogenic pathway for microarray data. Not only limit to the 31 oncogenic GSSs, our method can incorporate with other gene sets, such like the gene sets collected in MSigDB, to provide a systematic examining of the activities of pathways studied in previous literatures. The S-score values of gene sets can also incorporate with other clinical information, such like survival information and stage status, with different statistical methods for different proposes of studies in cancer. The flexibility of S-scare will be useful to uncover novel mechanism and pathway in cancer.
Supplementary Materials:
S1. S-Score performs and comparison with other scoring methods
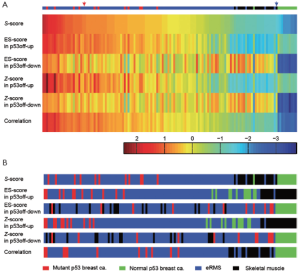
We first use the eRMS data, discussed in (34) to demonstrate the performance of the S-score, where we have applied S-score to classify different subtypes of sarcoma based on GSSs of p53 off, Ras on, Shh on, and Rb1 off (34). The classification has been validated by biological experiments and H&E staining. The promising results demonstrated the ability of S-score for scoring and clustering based on GSSs. Here we used one of the GSS, p53off, derived by comparing cancer samples with p53 mutation (marked in red in Figure 1) vs. samples with normal p53 (marked in green in Figure 1) to compare the capability of S-score against the other three scoring methods, ES-score (35), averaged Z-score (36), and Pearson correlation (37,38). The p53off GSS contains 150 up-regulated genes and 176 down-regulated genes (34,39). 112 expression profiles containing 94 embryonal rhabdomyosarcomas (eRMS) with unknown mutation status and 18 normal skeletal muscles (with normal p53) were applied for scoring (40). For the methods of ES-score and Z-score that can only process GSSs containing genes with the same direction of fold change (either up or down), up-regulated and down-regulated subsets of the p53off signature (p53off-up and p53off-down) were applied instead of whole signatures. The scores of the four scoring methods were shown in Figure S1A in descending order of S-score. Control samples (labeled as green), which were utilized to generate p53off signature, have the lowest scores in the methods of S-score, ES score in p53off-down, Z-score in p53off-down, and correlation. Normal skeletal muscle samples (labeled as black) have scores lower than the control samples in ES-score and Z-score of p53off-on. The scoring profiles of eRMS tumor samples, which are labeled as blue, have a similar index ranking pattern in S-score, ES-score in p53off-up, Z-score in p53off-up, and correlation. However, due to the limitation of ES-score and Z-score, the profiles of p53off-up and p53off-down subsets are quite different in both ES-score and Z-score methods. The order of the samples, which are sorted by the scores of each method, are shown in Figure S1B. The p53off scores of tumors are higher than most of the normal muscles in methods of S-score, ES-score in p53off-up, Z-score in p53off-up, and correlation. Notice that the distributions of tumor and normal muscles are mixed in p53off-down subset in both ES-score and Z-score. This result implicates that p53off-down subset has worse representation of the p53off status while they correctly assign index score to control samples. Clearly, without the integrated ability of both up- and down-regulated genes, it is difficult to obtain a conclusive score that faithfully reflects the expression pattern using ES-score and Z-score. With the capacity of integration, S-score and correlation-based method accurately project expression pattern to a signature score, with the suppression of the noisy effect of unrepresentative genes in the GSS, if they exist.
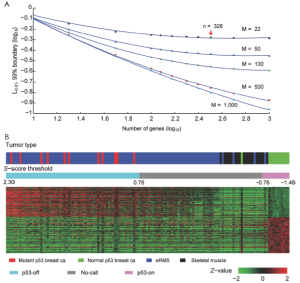
S3. Qualtitative status of signature
Although the S-score of each tumor sample was evaluated, the status of the signature (i.e., active or inactive) was not determined since simulations for the predictive interval need to be performed with current experiment setting. We have performed the simulation to cover a range of practical applications, and results are shown in Figure S5A. In the figure, we varied the number of genes in the signature set n=50 to 1,000, and the number of arrays M=22, 50, 100, 500, and 1,000. As expected, the boundaries are smaller with larger number of genes or more samples (chances of making “no call” shall be smaller with more genes in the gene set or more samples). For the parameters of p53off signature: n=326 and M=22, L0.01 was ±0.76, which is indicated by an arrow in Figure S5A. We will not make a status call for a given sample to be either “on” or “off” status of p53-off signature if the S-score is between (-0.76, 0.76) in order to guarantee no more than 1% of classification error. By applying the boundary value to the S-scores of test tumor samples, a total 52 of 97 eRMS samples (labeled as blue) were determined as p53-off status. Only two normal muscles and one eRMS (labeled as purple) were determined as p53-on status (Figure S5B).

Full table

Full table
Full table
Full table
Acknowledgments
Funding: This work was supported by the NIH/NCI cancer center grant (P30 CA054174-17), NIH/NCRR CTSA grant (1UL1RR025767) to YC, Cancer Prevention and Research Institute of Texas grant (CPRIT RP101195-C04) to GET, SC, and YC. TH is supported by the Greehey Children Cancer Research Institute (GCCRI) intramural research fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2012.12.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki. Institutional Review Board (IRB) and informed consent were not required since the data involved is from public sources.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ebi H, Tomida S, Takeuchi T, et al. Relationship of deregulated signaling converging onto mTOR with prognosis and classification of lung adenocarcinoma shown by two independent in silico analyses. Cancer Res 2009;69:4027-35. [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [PubMed]
- Gibbons DL, Lin W, Creighton CJ, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One 2009;4:e5401 [PubMed]
- Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology 2005;128:1655-67. [PubMed]
- Eckel F, Schmid RM. Chemotherapy in advanced biliary tract carcinoma: a pooled analysis of clinical trials. Br J Cancer 2007;96:896-902. [PubMed]
- Yusoff AR, Razak MM, Yoong BK, et al. Survival analysis of cholangiocarcinoma: a 10-year experience in Malaysia. World J Gastroenterol 2012;18:458-65. [PubMed]
- Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis 2004;24:115-25. [PubMed]
- Ohashi K, Nakajima Y, Kanehiro H, et al. Ki-ras mutations and p53 protein expressions in intrahepatic cholangiocarcinomas: relation to gross tumor morphology. Gastroenterology 1995;109:1612-7. [PubMed]
- Furubo S, Harada K, Shimonishi T, et al. Protein expression and genetic alterations of p53 and ras in intrahepatic cholangiocarcinoma. Histopathology 1999;35:230-40. [PubMed]
- Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut 2003;52:706-12. [PubMed]
- Rubin BP, Nishijo K, Chen HI, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell 2011;19:177-91. [PubMed]
- Mizuno H, Nakanishi Y, Ishii N, et al. A signature-based method for indexing cell cycle phase distribution from microarray profiles. BMC Genomics 2009;10:137. [PubMed]
- Cam H, Balciunaite E, Blais A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 2004;16:399-411. [PubMed]
- Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2004;2:E7 [PubMed]
- Andersen JB, Loi R, Perra A, et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology 2010;51:1401-9. [PubMed]
- Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 2008;68:1451-61. [PubMed]
- Lamb JR, Zhang C, Xie T, et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS One 2011;6:e20090 [PubMed]
- Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 2008;47:2059-67. [PubMed]
- Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 2008;3:e3088 [PubMed]
- Creighton CJ, Hilger AM, Murthy S, et al. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res 2006;66:3903-11. [PubMed]
- Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A 2010;107:6994-9. [PubMed]
- Jain R, Fischer S, Serra S, et al. The use of Cytokeratin 19 (CK19) immunohistochemistry in lesions of the pancreas, gastrointestinal tract, and liver. Appl Immunohistochem Mol Morphol 2010;18:9-15. [PubMed]
- Lie-A-Ling M, Bakker CT, Deurholt T, et al. Selection of tumour specific promoters for adenoviral gene therapy of cholangiocarcinoma. J Hepatol 2006;44:126-33. [PubMed]
- Balaton AJ, Nehama-Sibony M, Gotheil C, et al. Distinction between hepatocellular carcinoma, cholangiocarcinoma, and metastatic carcinoma based on immunohistochemical staining for carcinoembryonic antigen and for cytokeratin 19 on paraffin sections. J Pathol 1988;156:305-10. [PubMed]
- Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res 2010;70:3034-41. [PubMed]
- Argani P, Shaukat A, Kaushal M, et al. Differing rates of loss of DPC4 expression and of p53 overexpression among carcinomas of the proximal and distal bile ducts. Cancer 2001;91:1332-41. [PubMed]
- Tokumoto N, Ikeda S, Ishizaki Y, et al. Immunohistochemical and mutational analyses of Wnt signaling components and target genes in intrahepatic cholangiocarcinomas. Int J Oncol 2005;27:973-80. [PubMed]
- Bekaii-Saab T, Phelps MA, Li X, et al. Multi-institutional phase II study of selumetinib in patients with metastatic biliary cancers. J Clin Oncol 2011;29:2357-63. [PubMed]
- Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol 2012;57:69-76. [PubMed]
- Fava G. Molecular mechanisms of cholangiocarcinoma. World J Gastrointest Pathophysiol 2010;1:12-22. [PubMed]
- Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 2008;13:11-22. [PubMed]
- Andersen JB, Spee B, Blechacz BR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 2012;142:1021-1031.e15.
- Rubin BP, Nishijo K, Chen HI, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell 2011;19:177-91. [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [PubMed]
- Ebi H, Tomida S, Takeuchi T, et al. Relationship of deregulated signaling converging onto mTOR with prognosis and classification of lung adenocarcinoma shown by two independent in silico analyses. Cancer Res 2009;69:4027-35. [PubMed]
- Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A 2009;106:13820-5. [PubMed]
- Gibbons DL, Lin W, Creighton CJ, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One 2009;4:e5401 [PubMed]
- Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A 2005;102:13550-5. [PubMed]
- Laé M, Ahn EH, Mercado GE, et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol 2007;212:143-51. [PubMed]
- Cam H, Balciunaite E, Blais A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 2004;16:399-411. [PubMed]
- Cam H, Balciunaite E, Blais A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 2004;16:399-411. [PubMed]
- Andersen JB, Loi R, Perra A, et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology 2010;51:1401-9. [PubMed]
- Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 2008;68:1451-61. [PubMed]
- Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2004;2:E7 [PubMed]
- Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 2008;3:e3088 [PubMed]
- Creighton CJ, Hilger AM, Murthy S, et al. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res 2006;66:3903-11. [PubMed]
- Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009;15:489-500. [PubMed]
- Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 2008;47:2059-67. [PubMed]
- Lamb JR, Zhang C, Xie T, et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS One 2011;6:e20090 [PubMed]
- Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410-6. [PubMed]
- Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A 2010;107:6994-9. [PubMed]


