
A preliminary study of concurrent gains and losses across gene expression profiles and comparative genomic hybridization in Taiwanese breast cancer patients
Introduction
Breast cancer is heterogeneous in terms of molecular aberrations, and microarray experiments have revealed distinct molecular subtypes based on gene expression profiles, with some prognostic value (1-10). Breast cancer also displays genomic DNA copy number changes, although the complexity of genomic variations and the compromised resolution in conventional metaphase chromosome comparative genomic hybridization (CGH) have limited our understanding of genomic aberrations in this disease.
It was not until recently that microarray-based CGH (array CGH) experiments, either with BAC clones or synthesized oligonucleotide probes, provided an opportunity to evaluate the chromosomal instability of solid tumors other than cultured cells (11-13). It is believed that whole-genome array CGH can provide global insight into the fundamental processes of chromosomal instability that lead to breast cancer oncogenesis (14,15). It is supposed that cancer cells result from the progressive accumulation of genetic aberrations; amplified regions may contain dominant oncogenes whereas deleted regions may harbor tumor suppressor genes (16).
Because variations in genomic DNA copy number would have a direct impact on transcription due to complex regulation, the interplay between copy number changes and gene expression profiles deserves extensive evaluation. Genes displaying coherent patterns at the chromosomal and transcriptional levels are more likely to serve as potential predictive biomarkers for cancer therapy (17-20). We hypothesized that breast cancer tumorigenesis could originate at the chromosome level as DNA copy number changes and persist through mRNA transcription, manifesting in gene expression profiles.
In the current study, we performed genome-wide characterization of Taiwanese breast cancer by integrating 2 microarray technologies, array CGH and gene expression microarrays, to reveal genes with coherent patterns in both genomic and transcriptional aberrations in an effort to enhance our understanding of sporadic breast cancer and facilitate the discovery of potential biomarkers with therapeutic value.
Methods
Study population
Eligible patients were newly diagnosed with breast cancers and were scheduled for curative surgery (modified radical mastectomy or breast conservative therapy, both with axillary lymph node dissection) between January 2007 and December 2007. Informed consent was obtained pre-operatively, and the study protocol was reviewed and approved by the Institutional Review Board of Cathay General Hospital. Samples were snap frozen in liquid nitrogen and stored at -80 °C during surgery, and the relevant clinical data were retrieved from the cancer registry. Estrogen receptor (ER) positivity was defined as having at least 10% of nuclei stained positive through immunohistochemical (IHC) methods. On the other hand, ER negativity was claimed when none of the nuclei (0%) showed detectable IHC staining.
RNA extraction, reverse transcription, and expression arrays
Total RNA was extracted from frozen specimens by the TRIzol® reagent (Invitrogen, Carlsbad, CA). Purification of RNA was performed using RNeasy® mini kits (Qiagen, Valencia, CA), according to the manufacturer’s instructions. RNA integration was checked by gel electrophoresis; 2 bands of 18 and 28 s represented satisfactory RNA quality. Affymetrix GeneChip® Human Genome U133 plus 2.0 (Affymetrix, Santa Clara, CA) was used for the microarray experiments. Hybridization and scanning were performed according to the standard Affymetrix protocol. Image scanning was performed using a GeneChip® Scanner 3000, and scanned images were processed using the GeneChip® Operating Software (GCOS) and Affymetrix’s Microarray Suite (MAS) software to generate detection P values. Quantile normalization was used to normalize background-adjusted signals, and median polish was used for probeset normalization. The Robust Multichip Average (RMA) algorithm was applied for perfect match (PM) probe signals within the study (21). Probesets showing 2-fold changes in log2 ratio in either direction were selected for downstream analysis.
CGH microarrays
DNA was extracted using the QIAamp DNA® mini kit (Qiagen) from cancerous and matching normal breast tissue following RNA extraction. A minimal of 4 µg of total DNA was needed, and the purity and concentration of genomic DNA was verified using Bioanalyzer 2100® (Agilent, Santa Clara, CA). DNA quality control was indicated by an OD260/280 ratio greater than 1.8, according to the manufacturer’s instructions. The Agilent Human Genome 105k® microarray provided genome-wide coverage with an emphasis on the most commonly studied genomic coding regions and cancer-related genes. It included 99,000 probes that spanned the human genome with an average spatial resolution of approximately 15 kb, including coding and noncoding sequences. Genomic DNA and matching normal controls were labeled and hybridized to microarray slides for each study subject. After hybridization, the slides were scanned using the GeneChip® Scanner 3000 and the fluorescent dye ratios, which represented DNA copy number changes, were obtained for data analysis. The gene-focused content of the Agilent® array CGH facilitated the comparison of CGH and gene expression data so that we could correlate genomic copy number variations with gene expression patterns.
The analysis of CGH data began with the segmentation of normalized data, followed by identification of common (recurrent) gains and losses across multiple array CGH experiments. We used a cubic-spline curve fitting method to trace the ridgeline on 2D intensity distribution profiles, and used the Expectation-Maximization algorithm to accurately locate the dominant peak (22). Faster circular binary segmentation (FCBS), based on the DNAcopy algorithm, converted the normalized array CGH data into discrete segments of equal chromosomal copy number (23). After segmentation, we applied the value of derivative log-ratio spread (DLRS) from each array CGH experiment as a threshold to determine the gain or loss state of each segment.
Concurrent gains and losses
Concurrent gains and losses were detected from common probes across array CGH and gene expression experiments (53,670 common probes). We integrated gene expression and array CGH data to identify genes whose transcriptional levels were affected by changes in DNA copy number. Concurrent gains and losses were declared if and only if significant changes in a coherent manner were observed for both gene expression and array CGH platforms within the same study subject (Spearman’s correlation coefficient greater than 0.5 with Bonferroni correction of P-values less than 10-3). For subgroup analysis, the Cochran-Mantel-Haenszel test was used to identify concurrent genes displaying significant relevance to clinical ER status within each cytogenetic band (with Bonferroni correction of P-values less than 10-3).
Results
Global concurrent distributions
Of the 14 breast cancer samples that were assayed, 7 were ER-positive. As expected, breast cancer showed heterogeneous patterns in genomic variations and gene expression profiles. In global concurrent distributions, 12,022 probes showed concurrent gain or loss in at least one of the study subjects: 7,944 probes with concurrent gains (in 1-5 study samples) and 4,475 probes with concurrent losses (in 1-4 samples). In addition, 397 probes harbored both concurrent gains and losses, indicating a more complex gene expression/genomic variation interaction.
Repeated concurrences
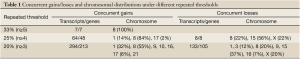
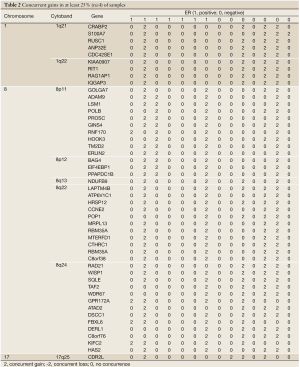
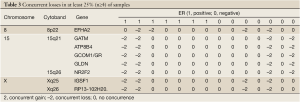
Varying numbers of concurrent genes were obtained with different thresholds of repeated concurrences (Table 1). When 25% of the samples (at least 4 samples showing coherent patterns) were requested, there were 48 genes showing concurrent gains and 9 genes with concurrent losses. 1q21-22, 8p11-12, 8q22, and 8q24 were the cytogenic bands harboring densely spaced concurrent gains, and concurrent losses were seen in 15q21 (Tables 2,3). When the threshold of repeated concurrences was relaxed to 20% (at least 3 subjects showing concurrence), genes with concurrent gains and losses were raised to 213 and 105, respectively. Table 1 summarized the numbers of repeated concurrences and chromosomal distributions under different thresholds. In the most stringent 33% of repeated concurrences, 7 concurrent gains were identified, namely LAPTM4B, HRSP12, WISP1, SQLE, GINS4, LYZ, and DSCC1, all of which were located in chromosome 8.
Full table
Full table
Full table
Subgroup analysis by ER
Subgroup comparison between 7 ER-positive and 7 ER-negative breast cancer samples was performed from genes displaying concurrent gains/losses in at least 3 (20%) of the assayed samples. 294 concurrent gains and 133 concurrent losses were observed, and these were reduced to 30 and 27 cytobands, respectively. The Cochran-Mantel-Haenszel test was used within each cytogenetic band (P-value <10-3 after Bonferroni correction). Concurrent gains were more common among ER-negative cancers in 1p32, 1p34, 1q21-23, and 17q25, whereas for ER-positive cancers, 8p11 showed repeated concurrent gain. In case of concurrent losses, 8p21 was significantly associated with the ER-positive phenotype.
Discussion
In the current study, we found that breast cancer was heterogeneous in case of both gene expression and variations in genomic DNA copy number. Concurrent gains were far more common than concurrent losses, and 7 candidate genes that displayed repeated concurrent gains in one-third of the assayed breast cancers were observed.
Nowadays, adjuvant therapy for breast cancer is based on certain established clinical prognostic factors and IHC results such as those for ER and human epidermal growth factor receptor 2 (HER2) over-expression. These parameters, however, are not sufficient for individualized therapy (24). In order to confront the heterogeneity not accounted for by conventional clinical and pathological factors, screening for potential biomarkers is one of the most urgent tasks in cancer therapy and genome medicine (25).
Chromosomal aberrations may induce gene expression variations. For instance, Zudaire et al. used traditional metaphase chromosome CGH to evaluate genomic aberrations associated with breast cancer and found that 16q loss was associated with better prognoses while 1q, 11q, 17q, and 20q gains were associated with poor prognoses (26). Nessling et al. used array CGH from 31 breast cancer samples with lymph node metastasis, and identified 37 gains and 13 losses from 112 candidate genes (27). Yau et al. found 2 molecular subtypes defined by array CGH in ER-positive breast cancers (28). There were also studies dealing with the correlations between copy number variations and gene expression profiles in breast cancer. Bergamaschi et al. analyzed array CGH results for 89 locally advanced breast cancers, of which gene expression profiles were used for molecular subtyping as defined by Stanford/UNC intrinsic signatures (4,20). They found that the basal-like subtype was associated with more gains/losses, while the luminal-B subtype had more frequent high-level DNA amplification. It should be noted that both basal-like and luminal-B subtypes were among the worst prognostic category in ER-negative and -positive breast cancers, respectively. Han et al. found several copy number gains (>25% in 28 samples) from triple-negative breast cancers and ascertained a concurrent gain in NF1B in the triple-negative phenotype (18). Chin et al. found candidate genes in high-level amplification, while Andre et al. performed unsupervised clustering of array CGH data (29,30).
The main drawback of the aforementioned studies was that gene expression data was retrieved from a publicly available microarray depository but not from the same subjects examined for copy number variations. The merit of the current study is that both array CGH and gene expression were assayed on the same subject, and the potential bias of individual variability was eliminated.
One of the most common concurrent gains was that of WNT1-inducible-signaling pathway protein 1 (WISP1), which is downstream of the WNT pathway and had been reported in colon cancer tumorigenesis (31). Another candidate gene, lysosomal-associated transmembrane protein 4B (LAPTM4B) has been reported as a prognostic factor in hepatocellular carcinoma and lung cancer (32-34). LAPTM4B was also associated with breast cancer chemoresistance (35). The roles of LAPTM4B and WISP1 in breast cancer tumorigenesis, however, remain inconclusive, but might provide further opportunities for breast cancer therapy from their close relationship with other human malignancies.
Several cytogenetic regions harboring concurrent gains or losses specific to distinct ER status were also revealed in our study. We found that both 17q25 and 1q23 gains were significantly associated with the ER-negative phenotype, while Bergamaschi et al. also reported a 1q23 gain in ER-negative breast cancer, as well as a 17q25 gain in basal-like breast cancers, most of which were also ER-negative (20). Han et al. showed that 1q21-23 and 17q25 gains were common in triple-negative breast cancer, 8p11 gain in ER-positive breast cancer, and 1p32 gain in ER-positive/HER2 over expressing breast cancer, and their findings were grossly in agreement with our concurrent analysis (18).
Insufficient sample size, especially in subgroup analysis, was a limitation of our study. In the current study, we did develop an analytical approach to analyze genes with coherent patterns in gene expression and copy number variations and identified 7 such concurrent gain genes in one-third of our assayed samples. Using genomic and transcriptional data obtained from the parallel analysis of CGH and gene expression microarrays of the same individual, false discoveries in finding breast cancer biomarkers were reduced. Chromosomal aberrations seemed to play a major role in regulating gene transcription. We hope the results of this preliminary study will facilitate the development of screening methods for breast cancer biomarker when more samples become available.
Acknowledgments
Funding: The work was supported in part by Cathay Medical Research Institute grant CGH-MR-9609 and National Taiwan University grant 95R0066-BM01-01 and 98HM0001.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.02.07). EYC serves as the Editor-in-Chief of Translational Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained pre-operatively, and the study protocol was reviewed and approved by the Institutional Review Board of Cathay General Hospital.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hedenfalk I, Duggan D, Chen Y, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med 2001;344:539-48. [PubMed]
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [PubMed]
- Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74. [PubMed]
- Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003;100:8418-23. [PubMed]
- Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 2006;7:96. [PubMed]
- Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 2006;355:560-9. [PubMed]
- Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A 2003;100:10393-8. [PubMed]
- van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530-6. [PubMed]
- Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817-26. [PubMed]
- Huang E, Cheng SH, Dressman H, et al. Gene expression predictors of breast cancer outcomes. Lancet 2003;361:1590-6. [PubMed]
- Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 1998;20:207-11. [PubMed]
- Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 1999;23:41-6. [PubMed]
- Oostlander AE, Meijer GA, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet 2004;66:488-95. [PubMed]
- van Beers EH, Nederlof PM. Array-CGH and breast cancer. Breast Cancer Res 2006;8:210. [PubMed]
- Reis-Filho JS, Simpson PT, Gale T, et al. The molecular genetics of breast cancer: the contribution of comparative genomic hybridization. Pathol Res Pract 2005;201:713-25. [PubMed]
- Fazeny-Dörner B, Piribauer M, Wenzel C, et al. Cytogenetic and comparative genomic hybridization findings in four cases of breast cancer after neoadjuvant chemotherapy. Cancer Genet Cytogenet 2003;146:161-6. [PubMed]
- Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 2002;62:6240-5. [PubMed]
- Han W, Jung EM, Cho J, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer 2008;47:490-9. [PubMed]
- Haverty PM, Fridlyand J, Li L, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer 2008;47:530-42. [PubMed]
- Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 2006;45:1033-40. [PubMed]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249-64. [PubMed]
- Chen HI, Hsu FH, Jiang Y, et al. A probe-density-based analysis method for array CGH data: simulation, normalization and centralization. Bioinformatics 2008;24:1749-56. [PubMed]
- Hsu FH, Chen HI, Tsai MH, et al. A model-based circular binary segmentation algorithm for the analysis of array CGH data. BMC Res Notes 2011;4:394. [PubMed]
- Weigelt B, Mackay A, A’hern R, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol 2010;11:339-49. [PubMed]
- Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst 2007;99:147-57. [PubMed]
- Zudaire I, Odero MD, Caballero C, et al. Genomic imbalances detected by comparative genomic hybridization are prognostic markers in invasive ductal breast carcinomas. Histopathology 2002;40:547-55. [PubMed]
- Nessling M, Richter K, Schwaenen C, et al. Candidate genes in breast cancer revealed by microarray-based comparative genomic hybridization of archived tissue. Cancer Res 2005;65:439-47. [PubMed]
- Yau C, Fedele V, Roydasgupta R, et al. Aging impacts transcriptomes but not genomes of hormone-dependent breast cancers. Breast Cancer Res 2007;9:R59. [PubMed]
- Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 2006;10:529-41. [PubMed]
- Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 2009;15:441-51. [PubMed]
- Pennica D, Swanson TA, Welsh JW, et al. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci U S A 1998;95:14717-22. [PubMed]
- Peng C, Zhou RL, Shao GZ, et al. Expression of lysosome-associated protein transmembrane 4B-35 in cancer and its correlation with the differentiation status of hepatocellular carcinoma. World J Gastroenterol 2005;11:2704-8. [PubMed]
- Kasper G, Vogel A, Klaman I, et al. The human LAPTM4b transcript is upregulated in various types of solid tumours and seems to play a dual functional role during tumour progression. Cancer Lett 2005;224:93-103. [PubMed]
- Yang H, Xiong F, Qi R, et al. LAPTM4B-35 is a novel prognostic factor of hepatocellular carcinoma. J Surg Oncol 2010;101:363-9. [PubMed]
- Li Y, Zou L, Li Q, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 2010;16:214-8. [PubMed]


