MDMX under stress: the MDMX-MDM2 complex as stress signals hub
Introduction
The evolutionary conservation of p53 across invertebrate and vertebrate species underscores its prominence in preserving cellular fitness and tissue function in multicellular organisms. p53 contributes to the maintenance of cellular homeostasis chiefly by controlling the expression of numerous genes in response to intra- and extra-cellular stress cues, hence representing a central element of the stress-response and cell fate decision process. In order to preserve their fitness in an energetically sensible way, cells have to constantly make cost-effective decisions that range between survival and death. To prevent and repair damages that can threaten their genome stability and the integrity of other essential structures, cells must sense cellular damage and promptly activate an appropriate response, such as cell cycle arrest, DNA repair or, if the damage is more severe, apoptosis. However, living cells are constantly exposed to an intrinsic level of stress caused by normal processes like cycling and division. Arresting the cell cycle or inducing programmed cell death in response to these transient and mild stresses would be energetically insensible. Therefore, to maintain their homeostasis and fitness, cells have to size their stress response to the perturbation that triggered it. Similarly, cells exposed to growth promoting stimuli need to be able to ignore fortuitous growth factors and undergo proliferation only in response to consistent mitogenic signals. In order to do this, one of the restriction points they have to bypass is represented by p53-controlled anti-proliferative genes, which need to be downregulated in order for the cells to commit to cell cycle progression (1). In this context, it is essential for the cells and especially for actively proliferating cells to modulate the function and stability of p53, given its role as guardian of the cellular homeostasis.
The two major essential negative regulators of p53 are MDMX and MDM2, two homolog proteins that control p53 activity and turnover, hence keeping it in check during normal cell cycling. Loss of MDMX and MDM2 leads to activation of p53 in most tissues, though only radiosensitive tissues such as spleen and thymus display the onset of p53-dependent apoptosis (2). Thus, MDM proteins probably evolved to buffer unwanted p53 activation (3). Highly proliferative cells, such as embryonic tissues or the proliferative compartments of the intestinal epithelium and of the adult brain, are generally more sensitive to p53 activation and therefore need an effective MDM proteins system to keep it in check (4,5). For example, MDMX and MDM2 are preferentially expressed in the subventricular zone of the adult brain, which is the putative site of adult neurogenesis, and where depletion of either MDM gene leads to p53-dependent apoptosis (2,6,7). A high frequency of over expression or amplification of MDMX and MDM2 is also observed in glioblastoma (8), which is derived from stem cells in the subventricular zone of the brain, underscoring the importance of MDM proteins in safeguarding the homeostasis of proliferating cells.
p53, MDMX and MDM2 form a highly dynamic regulatory node that contains positive and negative feedback loops (9). MDM2 can target p53 for degradation, but is also a p53 transcriptional target gene itself, creating a negative loop controlling p53 turnover. MDM2 protein stability and E3 ligase activity are increased by MDMX interaction, but MDMX can also be targeted for degradation by MDM2 upon DNA damage stress, when MDM2 activity is diverted from p53 to MDMX, generating another negative feedback loop that buffers MDM2 activity (10). This complex regulatory core ensures an effective buffering of p53 in non-stress conditions and its prompt activation under conditions of stress, i.e., in response to DNA damage (9). As the essential upstream modulator of p53, the MDMX-MDM2 complex integrates multiple intra- and extra-cellular signaling pathways regulating p53 response to perturbations of cellular homeostasis (9,11).
Since it harbors both a nuclear localization signal (NLS) and a nuclear export signal (NES), MDM2 can be found both in the nucleus and in the cytoplasm, whilst MDMX is predominantly cytoplasmic and depends on MDM2 binding to translocate into the nucleus. Given its localization, it is plausible to hypothesize that MDMX, rather than MDM2, is the first recipient of signaling cues directed towards the MDMX-MDM2 complex and aimed at either suppressing or activating p53. Although ultimately both MDM proteins are required for an effective modulation of p53, they have been shown to display non-overlapping functions, which could also include their role in mediating upstream signaling pathways targeted at the p53 core. We propose that MDMX represents a “signaling hub” transducing mitogenic signals as well as stress signals to the MDMX-MDM2 complex and therefore to p53. In a cancer context, our lab has shown that at least two receptor tyrosine kinases can inhibit p53 activation via modulation of the MDMX-MDM2 complex stability and, specifically, through post-translational modifications of MDMX (12,13). These and other studies (14) suggest that MDMX could be a promising and rather safe therapeutic target for pharmacological restoration of p53 in tumors harboring wild-type p53.
MDMX-MDM2 complex
Few year after its discovery, an unbiased screen for MDM2-interacting proteins identified MDMX as binding partner of MDM2. However, despite overexpression studies showing that MDMX could suppress p53 transcriptional function, the p53 community remained skeptical about its biological relevance in the regulation of p53. It was unclear why another protein would be required to keep p53 in check, when MDM2 could already do the job. Several structural studied stressed the importance of RING:RING interaction between MDMX and MDM2 for MDM2 stability and optimal E3 ligase activity (15,16). It was also reported that the binding of MDMX and MDM2 is required for the E3 ligase to target p53 (17). However, the key findings that led to the establishment of MDMX as a key player in the p53 regulatory core came from mouse genetic studies. Loss-of-function studies confirmed the role of MDMX an essential negative regulator of p53 during embryonic development. The embryonic lethality of the MDMX mouse model could be rescued by p53 knockout, confirming the functional dependency of the observed embryonic defect (18-20). MDMX or MDM2 knockout in neuronal progenitor cells and postmitotic cells displayed different phenotypes but, importantly, both phenotypes could be fully rescued by p53 ablation, showing that both MDMX and MDM2 are required for p53 inhibition and confirming that MDM2 cannot compensate for MDMX loss (2). However, loss of both MDMX and MDM2 did not cause any further increase in p53 levels, compared to MDM2 alone, leaving unclear whether MDMX could regulate p53 independently of MDM2.
The most convincing data in supporting the importance of the MDMX-MDM2 complex have been produced by Huang et al. (21) and Pant et al. (22), who investigated the role of MDMX and MDM2 during embryonic development. Pant’s model was an MDMX mutant mouse with a deletion of the RING domain, which prevented the interaction between MDMX and MDM2. Huang et al. adopted a less invasive genetic approach, using a knockin mouse model with a single base substitution in the MDMX RING domain, MDMXC462A. In this model, MDMX and MDM2 could not bind together to form a complex, but would otherwise retain their normal functions, including their ability to independently inhibit p53. Both models were shown to be embryonic lethal and, importantly, their lethality could be rescued by concomitant p53 knockout. These studies showed that MDMX and MDM2 function together as a complex and that their interaction is required for an effective control of p53.
Structure and function of MDMX
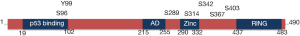
The MDMX protein shows a strong similarity to MDM2 and display four main conserved regions: (I) the amino-terminal hydrophobic region important for interaction with p53; (II) the carboxyl-terminal RING domain involved in heterodimerization with MDM2; (III) the zinc-finger domain and (IV) the acidic domain (AD) (Figure 1).
The p53 binding domain
The p53 binding domain is best conserved between MDM2 and MDMX. It interacts with the p53 transactivation domain (TAD) and inhibits its ability to induce transcription of its various targets (23,24). Although in vitro studies have reported similar affinities of the amino-terminal domains of MDMX and MDM2 for the p53 TAD (25), we should be careful in directly extrapolating these in vitro observations to live cells. For instance, Nutlin-3, which is a very potent inhibitor of MDM2-p53 interaction, is not very effective at inhibiting the MDMX-p53 interaction (26). This could be due to the fact that the p53-MDM protein interaction is affected by other domains within MDMX and MDM2. For instance, MDMX contains an internal motif, structurally similar to the interaction domain of p53, which can result in an internal interaction within MDMX protein, leading to diminished binding to p53. Furthermore, post-translational modifications or interactions with other proteins can modulate MDMX affinity with p53. For instance, MDMX p53-binding domain can be phosphorylated by c-Abl, at Tyr-99 and Tyr-55, interfering with p53 binding (27).
The RING domain
It is the second-best conserved domain between MDMX and MDM2. Whilst in MDM2 the RING-finger domain is essential for its ubiquitin ligase activity, allowing MDM2 to target p53 and other proteins for ubiquitination and degradation, in MDMX the RING domain does not display intrinsic ubiquitin ligase activity itself (28-30). Instead, the main functions of the RING domain of MDMX appear to be related to its interaction with MDM2. MDMX binding to MDM2 occurs via a RING:RING interaction that leads to the formation of a heterodimer that is both more stable and more effective in keeping p53 in check during embryonic development. Besides increasing MDM2 protein stability, MDMX biding can also stimulate its E3 ligase activity, providing an extended interaction domain for the E2 ubiquitin-conjugating enzyme (31,32).
The RING:RING interaction between MDMX and MDM2 is also required for the MDM2-mediated ubiquitination of MDMX upon stress, leading to a destabilization of the heterodimer and consequent activation of p53 (27-29). Post-translational modifications can affect this interaction, promoting ubiquitination by MDM2. Proteomic studies have identified several phosphorylation sites near the C-terminal RING domain of MDMX, including S342, S367 and S403. S342 and S367 are phosphorylated by Chk2, whilst S403 is modified by ATM. Phosphorylation of these sites promotes MDM2-targeting of MDMX for degradation, leading to decreased MDMX stability in response to DNA damage. Of note is that Ser367 is also phosphorylated by oncogenic kinase Akt, resulting in MDMX stabilization rather than degradation (33).
The MDMX RING domain also contains a cryptic NLS that is hidden by the intramolecular interaction. DNA damage can lead to phosphorylation of MDMX on several residues, including S367, which becomes a docking site for 14-3-3. The interaction of 14-3-3 with MDMX promotes a conformational change that disrupts the intramolecular interaction, exposes the cryptic NLS and leads to nuclear accumulation (34,35). Although on one side it may promotes MDMX degradation, since most MDM2 is in the nucleus, hence leading to p53 activation, it may on the other side be an active mechanism to suppress p53 activity.
The AD and zinc finger domain
Both MDM2 and MDMX contain an AD and zinc-finger domain, but their functionality is still poorly understood. MDM2 zinc-finger domain is involved in regulation of the retinoblastoma protein pRb (36), whereas the role of MDMX zinc-finger domain has not been fully elucidated, although also MDMX has been reported to interact with pRb (37,38).
MDM2 AD is required for its E3 ligase activity (29,30) and is involved in its interaction with p14ARF and ribosomal proteins. MDMX AD is involved in the intramolecular interactions with the p53-binding domain and the RING domain, but does not seem to interact with either p14ARF or ribosomal proteins, although p14ARF can affect the stability of MDMX through their interaction with MDM2Jeny (39-41).
The identified phosphorylation sites in MDMX (Table 1)

Full table
Phosphosites in the p53 binding domain: Tyr55, Tyr99 and Tyr96
The Tyr99 site is located in the p53 binding domain of MDMX and has been identified as a target phosphorylation site of the stress-activated kinase c-Abl. Upon DNA damage, the activation of c-Abl leads to the phosphorylation of MDMX, and also MDM2, causing p53 to be relieved from its major inhibitors and therefore activated (42). More recent modeling studies reveal that the phosphate group of pTyr99 imposes extensive steric clashes with the C-terminus of p53, contributing to the dramatic decrease in the binding affinity of MDMX for p53. This study shows how stress-induced MDMX phosphorylation dislodges p53 from the inhibitory complex and activates it in response to DNA damage (27). Another site that is phosphorylated by c-Abl upon DNA damage is the Tyr55. Phosphorylation on Tyr-99 is required at least partially for an efficient phosphorylation of Tyr-55. The Tyr96 site is phosphorylated by CDK2/p34, which is usually active in non-stress conditions, leading to an increase in the cytoplasmic localization of MDMX and promoting nuclear export of MDM2 (43).
Phosphosites in the AD and zinc domain: Ser289 and Ser314
The Ser289 site, when phosphorylated by its upstream kinase CK1A, has been shown to suppress MDMX interaction with p53, despite being located in the AD and not in the p53 binding domain. The ability of MDMX to inhibit the sequence-specific DNA binding activity of p53 requires the cooperation between MDMX and CK1α, and the phosphorylation of S289 on MDMX. A proteolytic fragment release assay revealed that in the MDMX-p53 complex, the MDMX AD and RING domain interact stably with the p53 DNA binding domain. These interactions are referred to as secondary interactions because they only occur after the canonical-specific binding between the MDMX and p53 N termini, but exhibit significant binding stability in the mature complex (44).
The Ser314 site has been recently identified by a phosphoproteomics screen as target of phosphorylation upon overexpression or ligand stimulation of the receptor tyrosine kinases Her4 and AXL, which were shown to promote its phosphorylation indirectly, through the mediation of the kinases CDK4/6 and p38. Phosphorylation of MDMX on Ser314 was able to favor MDMX nuclear import and increase its stability by protecting it from MDM2-mediated degradation (12,13). The identification of the Ser314 site as a target of multiple tyrosine kinases signaling point at a role of MDMX as the converging point, or “hub”, of oncogenic signaling aimed at suppressing p53, given the prosurvival role of tyrosine kinases in cancer development and the fact that p53 functional deactivation is required for tumorigenesis.
Phosphosites in the RING domain and proximal regions: Ser342, Ser367 and Ser403
Double strand-breaks (DSB) leads to ATM activation and phosphorylation of MDMX on three sites: S342, S367 and S403. S403 is a direct ATM target, whereas phosphorylation of S342 and S367 require the Chk2 kinase. Chk2 also stimulates MDMX ubiquitination and degradation by MDM2. Therefore, the E3 ligase activity of MDM2 is redirected to MDMX after DNA damage and contributes to p53 activation (45,46). Furthermore, DSB-induced phosphorylation of Ser-342 and Ser-367 was shown to be essential for creating a binding site for several isoforms of the 14-3-3 protein, leading to nuclear localization of MDMX, and promoting its degradation upon DNA damage (47). Another important component of the ATM-MDMX-p53 loop is the kinase Wip1 (or PPM1D), which contributes to p53 regulation by dephosphorylation of MDM2 and MDMX. The ATM-target Ser403 is directly dephosphorylated by Wip1, whereas Wip1 indirectly suppresses phosphorylation of MDMX on Ser342 and Ser367. In this way Wip1 inhibits the DNA damage-induced ubiquitination and degradation of MDMX, leading to the stabilization of MDMX and reduction of p53 activities (48). Another upstream kinase of Ser342 is AMPK, which is activated by metabolic stress. AMPK-mediated phosphorylation of MDMX leads to enhanced association between MDMX and 14-3-3, resulting in inhibition of p53 ubiquitination and p53 stabilization and activation (49).
MDMX as a stress signals hub
DNA damage
Whilst MDM2 undergoes self-ubiquitination and has a very short half-life in cell culture (15–30 min), MDMX half-life is much longer (3–6 hours) and its stability is controlled by MDM2-dependent ubiquitination in a stress-dependent manner (50). Following DNA damage by ionizing radiation, MDMX is rapidly degraded by MDM2, reliving p53 from the heterocomplex suppression and allowing its activation. Candidate approach and mass spectrometry have identified several phosphorylation sites (i.e., Ser342, Ser367 and Ser403) near the C-terminal RING domain of MDMX that are involved in MDMX degradation upon DNA damage (45,46). ATM phosphorylates MDMX at Ser403, causing MDM2 to target MDMX for ubiquitination. The Ser367 site has been shown to be phosphorylated upon ionizing radiation or DNA damaging treatment with camptothecin, leading to MDMX binding to 14-3-3 and translocating into the nucleus (34,35). Also, phosphorylation of MDMX at Tyr99 disrupt its interaction with p53, leading to p53 activation (14).
Ribosomal stress
MDMX has also been shown to be involved in the response to ribosomal stress. Ribosomal stress resulting from inhibition of rRNA transcription promotes MDMX degradation through L11-MDM2 interaction, which promotes MDM2-dependent ubiquitination of MDMX (39). Ribosomal subunit S7 has been shown to inhibit MDM2 and stabilize p53 in a MDMX-dependent fashion (51). Another study showed that the ribosomal noncoding 5S rRNA can stabilize MDMX under non-stress conditions (39).
Oncogenic stress
MDMX can also be modulated by oncogenic stress. ARF expression and interaction with MDM2 stimulates MDMX ubiquitination and degradation. K-Ras and insulin-like growth factor-1 (IGF-1) can increase MDMX transcript and protein levels (52). Another study by Li et al. showed that ARF can promote MDMX-MDM2 complex stability by mediating a second site interaction between MDM2 and MDMX (40). Also Akt was found to stabilize both MDM2 and MDMX, leading to stabilization of the heterocomplex and p53 suppression (33). These results underscore the importance of the MDMX-MDM2 heterocomplex as a common central hub of multiple stress signals aimed at modulating p53 function and stability.
MDMX-MDM2 complex as therapeutic target
Wild-type p53 expressing cancers have a high frequency of MDMX and/or MDM2 overexpression, either at protein or at transcript levels (3). For instance, in wild-type p53-expressing cutaneous melanoma, the frequency of MDM2 and MDMX protein alterations is 46% and 85%, respectively (3), second only to breast cancer, pointing at a major oncogenic role of MDM proteins in melanoma. However, Nutlin3 treatment in melanoma cells only caused modest reactivation of p53 (53,54), suggesting that targeting only MDM2-p53 interaction may not be sufficient to effectively restore p53 pathway. A specific inhibitor of the MDMX-p53 interaction has been reported, but its clinical application is limited by its complex mechanism of action (55,56). As an alternative, Bernal et al. developed a stabilized α-helical peptide, SAH-p53-8, able to bind MDMX and disrupt the interaction between MDMX and p53 in vitro and in vivo (57). Gembarska et al. (14) demonstrated that this peptide could induce p53-dependent apoptosis in cultured melanoma cells and, importantly, sensitize melanoma cells to conventional chemotherapy and BRAF-V600E inhibitors. One limitation of this approach, however, is the p53-independent oncogenic function of MDMX (58,59). An alternative approach that would overcome this limitation is targeting MDMX protein abundance by using alternative splice-switching antisense oligonucleotides (ASOs) of exon-intron boundaries of exon 6. Treatment of cultured melanoma cells to these agents led to exon 6 skipping and decrease of MDMX protein abundance, resulting in increased p53 activity (60). In vivo, these agents were able to suppress the growth of patient-derived xenografts (PDX) and sensitize them to MAPK-targeting inhibitors. Although these results are very encouraging, genetic studies (21,22) have shown that MDMX and MDM2 exert their control on p53 more efficiently when working together as a complex, rather than singularly. These observations suggest that targeting the stability of the MDMX-MDM2 complex could potentially provide a more effective strategy to rescue p53 than therapies targeted only on MDMX or MDM2. For instance, the design of peptides able to bind MDMX and disrupt its interaction with MDM2 could potentially prove more effective, although these peptides are expensive to produce and have to be delivered intravenously by trained professionals, making their use in the clinic logistically difficult. An alternative method of disassociating the MDMX-MDM2 complex needs to be explored for therapeutics.
Acknowledgments
Funding: This work was supported in part by the Morningside Foundation, the Zhu fund and grants from NIH/NCI (R01CA085679, RO1CA167814, and RO1CA183074).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “p53 Biology and Cancer”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.18). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. ZMY served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Translational Cancer Research. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zwang Y, Sas-Chen A, Drier Y, et al. Two Phases of Mitogenic Signaling Unveil Roles for p53 and EGR1 in Elimination of Inconsistent Growth Signals. Mol Cell 2011;42:524-35. [Crossref] [PubMed]
- Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA 2006;103:3232-7. [Crossref] [PubMed]
- Wade M, Li Y, Wahl G. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013;13:83-96. [Crossref] [PubMed]
- Valentin-Vega YA, Box N, Terzian T, et al. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 2009;77:442-9. [Crossref] [PubMed]
- Marine JC. MDMX (MDM4), a Promising Target for p53 Reactivation Therapy and Beyond. Cold Spring Harb Perspect Med 2016;6:a026237 [Crossref] [PubMed]
- Ringshausen I, O’Shea CC, Finch AJ, et al. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 2006;10:501-14. [Crossref] [PubMed]
- Garcia D, Warr MR, Martins CP, et al. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev 2011;25:1746-57. [Crossref] [PubMed]
- Rao SK, Edwards J, Joshi A, et al. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol 2010;96:169-79. [Crossref] [PubMed]
- Shadfan M, Lopez-Pajares V, Yuan ZM. MDM2 and MDMX: Alone and together in regulation of p53. Transl Cancer Res 2012;1:88-9. [PubMed]
- Chen J. The roles of MDM2 and MDMX phosphorylation in stress signaling to p53. Genes Cancer 2012;3:274-82. [Crossref] [PubMed]
- Li J, Kurokawa M. Regulation of MDM2 stability after DNA damage. J Cell Physiol 2015;230:2318-27. [Crossref] [PubMed]
- Gerarduzzi C, de Polo A, Liu XS, et al. Human epidermal growth factor receptor 4 (Her4) Suppresses p53 Protein via Targeting the MDMX-MDM2 Protein Complex: IMPLICATION OF A NOVEL MDMX SER-314 PHOSPHOSITE. J Biol Chem 2016;291:25937-49. [Crossref] [PubMed]
- de Polo A, Luo Z, Gerarduzzi C, et al. AXL receptor signaling suppresses p53 in melanoma through stabilization of the MDMX-MDM2 complex. J Mol Cell Biol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Gembarska A, Luciani F, Fedele C, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nature Med 2012;18:1239-47. [Crossref] [PubMed]
- Kawai H, Lopez-Pajares V, Kim MM, et al. RING Domain–Mediated Interaction Is a Requirement for MDM2's E3 Ligase Activity. Cancer Res 2007;67:6026-30. [Crossref] [PubMed]
- Linares LK, Hengstermann A, Ciechanover A, et al. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci 2003;100:12009-14. [Crossref] [PubMed]
- Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett 2009;583:2710-4. [Crossref] [PubMed]
- Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29:92-5. [Crossref] [PubMed]
- Finch RA, Donoviel DB, Potter D, et al. MDMX is a negative regulator of p53 activity in vivo. Cancer Res 2002;62:3221-5. [PubMed]
- Migliorini D, Denchi EL, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 2002;22:5527-38. [Crossref] [PubMed]
- Huang L, Yan Z, Liao X, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci USA 2011;108:12001-6. [Crossref] [PubMed]
- Pant V, Xiong S, Iwakuma T, et al. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA 2011;108:11995-2000. [Crossref] [PubMed]
- Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 1996;15:5349. [PubMed]
- Shvarts A, Bazuine M, Dekker P, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 1997;43:34-42. [Crossref] [PubMed]
- Böttger V, Bottger A, Garcia-Echeverria C, et al. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene 1999;18:189-99. [Crossref] [PubMed]
- Joseph TL, Madhumalar A, Brown CJ, et al. Differential binding of p53 and nutlin to MDM2 and MDMX: Computational studies. Cell Cycle 2010;9:1167-81. [Crossref] [PubMed]
- Chen X, Gohain N, Zhan C, et al. Structural basis of how stress-induced MDMX phosphorylation activates p53. Oncogene 2016;35:1919-25. [Crossref] [PubMed]
- Kawai H, Wiederschain D, Kitao H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 2003;278:45946-53. [Crossref] [PubMed]
- Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol 2003;23:4939-47. [Crossref] [PubMed]
- Meulmeester E, Frenk R, Stad R, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol 2003;23:4929-38. [Crossref] [PubMed]
- Poyurovsky MV, Priest C, Kentsis A, et al. The Mdm2 RING domain C—terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 2007;26:90-101. [Crossref] [PubMed]
- Linke K, Mace PD, Smith CA, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 2008;15:841-8. [Crossref] [PubMed]
- Lopez-Pajares V, Kim MM, Yuan ZM. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem 2008;283:13707-13. [Crossref] [PubMed]
- Jin Y, Dai MS, Lu SZ, et al. 14-3-3γ binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J 2006;25:1207-18. [Crossref] [PubMed]
- LeBron C, Chen L, Gilkes DM, et al. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J 2006;25:1196-206. [Crossref] [PubMed]
- Hsieh JK, Chan FS, O’Connor DJ, et al. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell 1999;3:181-93. [Crossref] [PubMed]
- Uchida C, Miwa S, Isobe T, et al. Effects of MdmX on Mdm2—mediated downregulation of pRB. FEBS Lett 2006;580:1753-8. [Crossref] [PubMed]
- Zhang H, Hu L, Qiu W, et al. MDMX exerts its oncogenic activity via suppression of retinoblastoma protein. Oncogene 2015;34:5560-9. [Crossref] [PubMed]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J 2006;25:5614-25. [Crossref] [PubMed]
- Li X, Gilkes D, Li B, et al. Abnormal MDMX degradation in tumor cells due to ARF deficiency. Oncogene 2012;31:3721-32. [Crossref] [PubMed]
- Daftuar L, Zhu Y, Jacq X, et al. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE 2013;8:e68667 [Crossref] [PubMed]
- Zuckerman V, Lenos K, Popowicz GM, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 2009;284:4031-9. [Crossref] [PubMed]
- Elias B, Laine A, Ronai ZE. Phosphorylation of MdmX by CDK2/Cdc2p34 is required for nuclear export of Mdm2. Oncogene 2005;24:2574-9. [Crossref] [PubMed]
- Wei X, Wu S, Song T, et al. Secondary interaction between MDMX and p53 core domain inhibits p53 DNA binding. Proc Natl Acad Sci 2016;113:E2558-63. [Crossref] [PubMed]
- Pereg Y, Shkedy D, de Graaf P, et al. Phosphorylation of Hdmx mediates its Hdm2-and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA 2005;102:5056-61. [Crossref] [PubMed]
- Chen L, Gilkes DM, Pan Y, et al. ATM and Chk2−dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 2005;24:3411-22. [Crossref] [PubMed]
- Okamoto K, Kashima K, Pereg Y, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 2005;25:9608-20. [Crossref] [PubMed]
- Zhang X, Lin L, Guo H, et al. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res 2009;69:7960-8. [Crossref] [PubMed]
- He G, Zhang YW, Lee JH, et al. AMP-activated protein kinase induces p53 by phosphorylating MDMX and inhibiting its activity. Mol Cell Biol 2014;34:148-57. [Crossref] [PubMed]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol 2003;23:5113-21. [Crossref] [PubMed]
- Zhu Y, Poyurovsky MV, Li Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 2009;35:316-26. [Crossref] [PubMed]
- Gilkes DM, Pan Y, Coppola D, et al. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol 2008;28:1999-2010. [Crossref] [PubMed]
- de Lange J, Ly LV, Lodder K, et al. Synergistic growth inhibition based on small molecule p53 activation as treatment for intraocular melanoma. Oncogene 2012;31:1105-16. [Crossref] [PubMed]
- Ji Z, Njauw CN, Taylor M, et al. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibiton. J Invest Dermatol 2012;132:356-64. [Crossref] [PubMed]
- Reed D, Shen Y, Shelat AA, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem 2010;285:10786-96. [Crossref] [PubMed]
- Bista M, Smithson D, Pecak A, et al. On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53. Plos One 2012;7:e37518 [Crossref] [PubMed]
- Bernal F, Wade M, Godes M, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell 2010;18:411-22. [Crossref] [PubMed]
- de Lange J, Teunisse AF, Vries MV, et al. High levels of Hdmx promote cell growth in a subset of uveal melanomas. Am J Cancer Res 2012;2:492-507. [PubMed]
- Carrillo AM, Bouska A, Arrate MP, et al. Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene 2015;34:846-56. [Crossref] [PubMed]
- Dewaele M, Tabaglio T, Willekens K, et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 2016;126:68-84. [Crossref] [PubMed]