
KDM1 links nuclear GSK3β to epigenetic alterations
Accumulating evidence indicates that cancer stem cells (CSCs) play an important role during the processes of tumor development, progression, and recurrence and are primarily responsible for therapeutic resistance and poor clinical outcome of patients (1). Epigenetic alterations, particularly histone methylation, have been increasingly recognized as a global transcriptional regulator that contributes to stem cell self-renewal and differentiation under physiological and pathological conditions (2). For example, methylations of Lys9 and Lys27 residues of histone H3 (H3K9me2/3 and H3K27me3), associate with heterochromatin and transcriptional repression; whereas H3K4me2/3, often found in active gene promoters, is associated with transcriptional activation. These epigenetic modifications create unique promoter architectures that control gene expression. Glycogen synthase kinase 3 beta (GSK3β) has been shown to take part in the regulation of histone methylation including H3K4 methylation in the promoter regions of multiple genes (3,4). However, the molecular mechanisms for GSK3β in mediating alterations of histone methylation remain to be defined. In September 2016 issue of Nature Cell Biology, Zhou et al. (5) addressed this challenge and singled out a connection between nuclear GSK3β and epigenetic aberration via regulation of lysine-specific histone demethylase 1A (KDM1A) stability.
Histone lysine methylation is a dynamic and reversible process. KDM1A (also known as LSD1) is the first identified lysine-specific histone demethylase that selectively remove the methyl group from H3K4me1/2 through flavin-adenine-dinucleotide (FAD)-dependent oxidative reaction (6). KDM1A exerts its gene repression functions by forming as a key catalytic component in several corepressor complexes, including Co-REST, NuRD, CtBP, HDAC, or SIRT1 (7,8). KDM1A is often overexpressed in breast (9), bladder, lung, colon (10) cancers and glioblastoma (11), and is required for the maintenance and differentiation of stem cells (12) and CSCs (13). Despite its determinant roles in stem cell pluripotency and differentiation and tumorigenesis, the mechanisms that lead to KDM1A dysregulation in these tumors remain to be better delineated.
Consistent with the notion that KDM1A is a labile protein that under constant protein ubiquitination and degradation (6,9), Zhou et al. found that nuclear GSK3β stabilizes KDM1A by decreasing its ubiquitylation. GSK3β prefers substrates that are primed phosphorylated by another kinase at a Serine or Threonine residue located 4 or 5 amino acids downstream or upstream of GSK3β phosphorylation site (14). The authors found that KDM1A contains several potential GSK3β phosphorylation sites harboring a canonical –S/TxxxS/T-motif. They further identified that Ser687 of KDM1A is phosphorylated by casein kinase 1 alpha (CK1α) and this priming phosphorylation facilitates GSK3β phosphorylation at Ser683. As GSK3β does not directly deubiquitylate KDM1A, the authors proposed that there should be deubiquitylase (DUB) to cooperate the deubiquitylation of KDM1A. Thus, they screened a panel of DUBs and identified ubiquitin-specific protease 22 (USP22) as a direct DUB for KDM1A. Subsequently, the authors explored the roles of GSK3β-USP22-KDM1A axis in glioma. They showed that GSK3β-USP22-KDM1A axis is required for tumorigenesis of CSCs and associated with the grade of human glioma malignance.
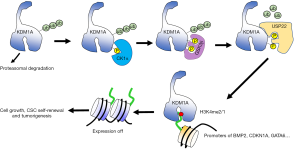
A picture appears in which KDM1A connects nuclear GSK3β with epigenetics (Figure 1). First, KDM1A is phosphorylated by CK1α at Ser687 which facilitates its binding with and phosphorylation by GSK3β. Subsequently, USP22, a direct KDM1A DUB, deubiquitylates KDM1A. Consequently, stabilized KDM1A removes the activation marks on H3K4 in promoter regions of stem cell pluripotency inhibition genes, including BMP2, CDKN1A and GATA6. Finally, as reported here, silencing of these tumor suppressors promotes tumorigenesis of CSCs. More importantly, the authors also showed therapeutic potential of tideglusib, a selective non-ATP-competitive GSK3 inhibitor with good blood-brain barrier penetration and now in a phase II trial for the treatment of Alzheimer’s disease, along or in combination with temozolomide, a FDA approved drug for glioma. Although accumulating numbers of oncogenes have been extensively characterized, many of these oncogenes are not good drug targets. Thus, there are urgent needs for alternative therapeutic approaches to interfere these oncogenes. As KDM1A is structurally related monoamine oxidases A & B (MAO-A and MAO-B), a KDM1A inhibitor may exhibit side-effect by suppressing MAO-A and MAO-B. In addition, blood-brain barrier makes it harder for KDM1A as a therapeutic target for treating glioma. Thus, targeting the proposed GSK3β-USP22-KDM1A signaling axis represents an excellent approach in suppressing the function of KDM1A in glioma. Recent works suggest that onco-DUBs, which contribute substantially to the stability of onco-proteins and pathways and are amenable to pharmacologic inhibition by small molecules, possess great potential for cancer treatment (15). Though not explored in their work, USP22, may also serve as a promising target for treating glioma and deserve further investigation. Nevertheless, the therapeutic potential established in this outstanding study will warrant further translational investigation and inspire many areas of basic inquiry that will further our understanding of the concerted connections between aberrant cell signaling and epigenetics during malignant tumor progression.
Acknowledgments
Funding: This work was supported by the Shared Resources of the University of Kentucky Markey Cancer Center (P30CA177558), grants from NIH (CA125454 and CA188118), DoD (BC140733P1), and Mary Kay Ash Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chen Qian (Center for Inflammation & Epigenetics, Houston Methodist Hospital Research Institute, Houston, TX, USA).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.26). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nguyen LV, Vanner R, Dirks P, et al. Cancer stem cells: an evolving concept. Nat Rev Cancer 2012;12:133-43. [PubMed]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 2007;8:307-18. [Crossref] [PubMed]
- Hill EV, Ng TH, Burton BR, et al. Glycogen synthase kinase-3 controls IL-10 expression in CD4(+) effector T-cell subsets through epigenetic modification of the IL-10 promoter. Eur J Immunol 2015;45:1103-15. [Crossref] [PubMed]
- Ougolkov AV, Bone ND, Fernandez-Zapico ME, et al. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukemia B cells. Blood 2007;110:735-42. [Crossref] [PubMed]
- Zhou A, Lin K, Zhang S, et al. Nuclear GSK3β promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat Cell Biol 2016;18:954-66. [Crossref] [PubMed]
- Shi YJ, Matson C, Lan F, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005;19:857-64. [Crossref] [PubMed]
- Mulligan P, Yang F, Di Stefano L, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 2011;42:689-99. [Crossref] [PubMed]
- Wang Y, Zhang H, Chen Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009;138:660-72. [Crossref] [PubMed]
- Wu Y, Wang Y, Yang XH, et al. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep 2013;5:224-36. [Crossref] [PubMed]
- Hayami S, Kelly JD, Cho HS, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer 2011;128:574-86. [Crossref] [PubMed]
- Sareddy GR, Nair BC, Krishnan SK, et al. KDM1 is a novel therapeutic target for the treatment of gliomas. Oncotarget 2013;4:18-28. [PubMed]
- Wang J, Hevi S, Kurash JK, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 2009;41:125-29. [Crossref] [PubMed]
- Wang J, Lu F, Ren Q, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res 2011;71:7238-49. [Crossref] [PubMed]
- Kaidanovich-Beilin O, Woodgett JR. GSK-3: Functional Insights from Cell Biology and Animal Models. Front Mol Neurosci 2011;4:40. [Crossref] [PubMed]
- Potu H, Peterson LF, Pal A, et al. Usp5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget 2014;5:5559-69. [Crossref] [PubMed]


