
GPD1L is down-regulated by a three-microRNA signature in pancreatic cancer
Introduction
Pancreatic cancer is the seventh most common cause of cancer deaths, resulting in 330,000 deaths annually around the world (1). Pancreatic ductal adenocarcinoma (PDAC) often has a very poor prognosis and the 5-year survival rate is only 5% (2). This fact urges comprehensive investigation of the molecular pathogenesis of PDAC. Recent studies have shown that microRNAs play important roles in different types of cancer including PDAC (3,4). The microRNAs could inhibit target gene expression by binding the complementary sequence within the 3'UTR of target genes (5). Oncogenes and tumor suppressor genes are vital targets for microRNAs in cancer (6).
Several studies have revealed distinct microRNA signatures in PDAC (7-13), but the target validation is often beyond the scope of these studies. On one hand, there appears to be a small overlap of highlighted microRNAs among different studies. The microRNA signature which could be cross-validated may have true biological relevance to the pathogenesis of PDAC. On the other hand, how the microRNA signatures affect their target genes and therefore the tumor phenotypes remains largely unknown. It is difficult to translate the microRNA signature into clinical benefits unless their target genes are validated (3).
In the present study, we compared the microRNA signatures from independent studies and identified a panel of five microRNAs which were reported to be reproducible. We measured the 5-microRNA panel in PDAC and validated an up-regulation of 3-microRNA signature (miR155, miR181a and miR221). The miRDB (14) was used to predict the targets of miR155, miR181a and miR221, respectively. We find that four genes (CREBRF, GABRA1, GPD1L and REPS2) have potential binding sites for all three microRNAs. We measured the mRNA levels of these four genes and found that GPD1L was down-regulated in PDAC. In addition, GPD1L down-regulation was associated with poorer prognosis of PDAC in three independent datasets from public databases. Further results from PDAC cell lines suggest that the 3-microRNA signature inhibited GPD1L expression through binding its 3'UTR and GPD1L reduction promoted aggressive tumor phenotypes. Our study identifies GPD1L as a novel target gene for a 3-microRNA signature and GPD1L might be an important therapeutic target in PDAC.
Methods
Clinical samples
The study was approved by the Institutional Review Boards of the first affiliated hospital of Wenzhou Medical University (Wenzhou, China) and written informed consent was obtained from each patient. Twenty pairs of PDAC tissues and adjacent normal tissues were collected in Department of Hepatobiliary Surgery, The First Affiliated Hospital of Wenzhou Medical University. Tissues were freshly resected during surgery and immediately frozen in liquid nitrogen for subsequent total RNA extraction.
Cell cultures
BxPC3 (CRL-1687) and PANC1 (CRL-1469) cell lines from ATCC were maintained in RPMI-1640 or DMEM with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 mg/mL streptomycin, respectively. Cells were cultured in a humidified atmosphere with 5% CO2 at 37 °C.
Cell proliferation assay
Cells transfected with GPD1L siRNA or control siRNA were seeded into a 96-well plate in triplicate at the concentration of 4×103 cells per well. The cell growth was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) bromide assay every 24 hours. Cells were incubated with 5 mg/mL MTT for 4 h, and subsequently solubilized in DMSO (100 µL/well). The absorbance at 570 nm was then measured using an ELISA reader.
Colony formation assay
Cells transfected with GPD1L siRNA or control siRNA were plated in duplicate in a 6-well plate. After incubation at 37 °C for 14 days, the colonies were stained with crystal violet solution in methanol for 15 min. Colonies >50 µm in diameter were counted.
Real-time PCR
To detect the relative levels of microRNA panel and their target genes (CREBRF, GABRA1, GPD1L and REPS2), quantitative real time-PCR (qPCR) was performed. Briefly, Total RNA was isolated from tissues or cell lines using TRIzol reagent, according the manufacture’s protocol. For CREBRF, GABRA1, GPD1L and REPS2, the cDNA was generated through reverse transcription using MMLV reverse transcriptases (Promega) and actin was used as an endogenous control. The qRT-PCR primers were as follows. CREBRF: ACCCACTTCAAGCACACAAAT (F), GGGTTGATCTTTACCTTTGCCT (R); GABRA1: AGCCGTCATTACAAGATGAACTT (F), TGGTCTCAGGCGATTGTCATAA (R); GPD1L: GTTGCCATGTCAAATCTTAGCG (F), GCACTCTCCCAGTGATCTCAT (R) and REPS2: AGGAAACACAGTCTCCCACGA (F) and CTCCGCAGTTTGCTGTAGC (R); actin: CGCAGCCACTGTCGAGTC (F) and CGGAGTCCATCACAATGCCT (R). For mature microRNA quantification, cDNA was synthesized using specific stem-loop universal primers (60 ng) and a TaqMan microRNA reverse transcription kit. U6 small nuclear RNA was used as an internal control. The reaction condition was as follows: 30°C for 10 min; 42 °C for 1 h; 85 °C for 5 min; 5 °C for 5 min. TaqMan miRNA assays for miR-100 (000437), miR-146a (000468), miR-155 (002623), miR-181a (000480), miR-221 (000524) and U6 snRNA (001973) were from Applied Biosystems. The qPCR conditions were 95 °C for 2 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The fold change for each gene relative to the control group was calculated using the ΔΔCt method. The miR mimics and negative control (NC) were from Ambion.
Luciferase activity assay
To construct the 3'UTR luciferase vector of GPD1L containing binding sites for miR-155, miR-181a and miR-221, 2,500 bp of the 3'UTR sequence of the human GPD1L was amplified by PCR and cloned into pMIR-report. For luciferase reporter assay, BxPC3 and PANC1 cells were transiently transfected with pMIR-report containing GPD1L 3'-UTR and miR mimics using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, reporter gene activity was measured using the dual-luciferase assay-system (Promega). Renilla luciferase activity was used to normalize for transfection efficiency.
Annexin V-FITC apoptosis assay
Cell lines were transfected with GPD1L siRNA or control siRNA for 2 days and stained with Annexin V-FITC Apoptosis kit (abcam, ab14085) according to the manufacturer’s instruction. Then, the stained cells were analyzed by MoFlo XDP (Beckman Coulter, Inc.).
Western blot
Proteins were extracted from cell lines with RIPA buffer. Protein samples were resolved by SDS-PAGE and analyzed by immunoblots. GPD1L antibody was from Abcam (ab107509) and used at 1:500 dilution.
Statistical analysis
All data from clinical samples were presented as whiskers-box plots and differences between two groups were calculated with the nonparametric Mann Whitney U test with Bonferroni correction. All data from cell lines were presented as mean with SD and differences between two groups were calculated with two-tailed Student t test. Statistical analysis was performed using GraphPad Prism. Statistically significant differences were defined as P<0.05. For all, *, P<0.05; **, P<0.01; ***, P<0.001.
Results
Validation of microRNA signature in PDAC
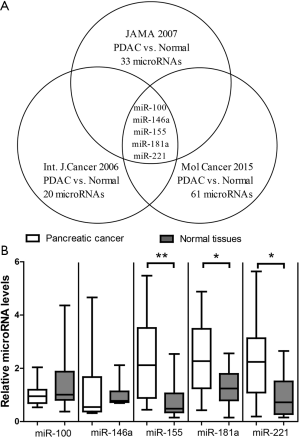
We first compared the microRNA signatures from previous studies. Bloomston and colleagues used microRNA microarray and reported that 33 microRNAs were differentially expressed between pancreatic cancer and normal tissues (7). Lee and colleagues used quantitative real-time PCR assay and reported that 20 microRNAs were differentially expressed between pancreatic cancer and normal tissues (8). Muller and colleagues used next-generation sequencing and reported that 61 microRNAs were differentially expressed between pancreatic cancer and normal tissues (12). Among these three lists, 5-microRNAs including miR-100, miR-146a, miR-155, miR-181a, miR-221 appears to be reproducible across different technical platforms (Figure 1A). The relatively small overlap of highlighted microRNAs in these three highly cited studies suggests that it is necessary to independently validate the 5-microRNA panel. Thus, we measured the 5-microRNA panel in PDAC. The results of TaqMan assays showed that miR-155, miR-181a and miR-221 were significantly up-regulated in PDAC after Bonferroni correction for multiple tests (Figure 1B). In contrast, there was no significant difference for other two microRNAs. As the 3-microRNA signature consisting of miR-155, miR-181a and miR-221 could be well cross-validated, they may have true biological relevance to PDAC and their target genes may have important roles in the pathogenesis of PDAC.
Target genes of the 3-microRNA signature in PDAC
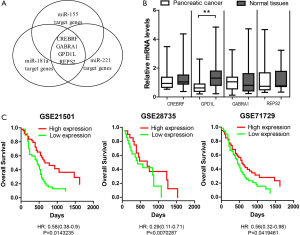
Next, we tried to identify the target genes of the 3-microRNA signature. Although each microRNA has numerous target genes, only few of them truly contribute to the tumor phenotypes. Thus, we hypothesize that the genes targeted by all three microRNAs are critical mediators for the 3-microRNA signature to affect tumor phenotypes. The miRDB was used to predict the targets of miR155, miR181a and miR221, respectively. We find that four genes (CREBRF, GABRA1, GPD1L and REPS2) have potential binding sites for all three microRNAs (Figure 2A). It suggests that these four genes are more likely to be down-regulated by the 3-microRNA signature in PDAC. We measured the mRNA levels of these four genes and found that GPD1L was down-regulated after Bonferroni correction (Figure 2B). To assess whether GPD1L plays an important role in PDAC, we analyzed the effects of GPD1L expression on patient survival in four independent open-access datasets [GSE21501 (15), GSE28735 (16), GSE71729 (17) and GSE57495 (18)] through PROGgeneV2 Database (19). The patients were classified as GPD1L high-expression or low-expression groups and their Kaplan-Meier survival curves were compared (Figure 2C). The results showed that the patients with high GPD1L expression had significantly longer survival time than those with low GPD1L expression in three out of four dataset: GSE21501 (HR =0.58; P=0.0143235), GSE28735 (HR =0.29; P=0.0070287), GSE71729 (HR =0.56; P=0.0419461) and GSE57495 (HR =0.97; P=0.91). It indicates that GPD1L down-regulation is strongly associated with poorer prognosis of PDAC.
GPD1L is a direct target of the 3-microRNA signature
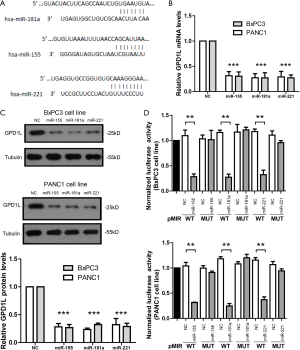
To further confirm if GPD1L is a direct target of the 3-microRNA signature (Figure 3A), we over-expressed miR155, miR181a or miR221 mimics in BxPC3 and PANC1 cell lines, respectively. Results of real time-PCR (Figure 3B) and WB (Figure 3C) showed that miR155, miR181a or miR221 mimics could inhibit the mRNA and protein levels of GPD1L, respectively. Next, we cloned GPD1L 3'UTR containing binding sites of miR155, miR181a and miR221 into a luciferase reporter vector pMIR. When BxPC3 and PANC1 cells were transfected with luciferase reporter vector and miR155, miR181a or miR221 mimics, the luciferase activity was greatly reduced (Figure 3D). Once the predicted binding sites in the 3'UTR were mutated, the inhibitory effects of miR155, miR181a or miR221 mimics were abolished. It indicates that miR155, miR181a or miR221 could directly bind to GPD1L 3'UTR.
GPD1L RNAi promotes aggressive tumor phenotypes
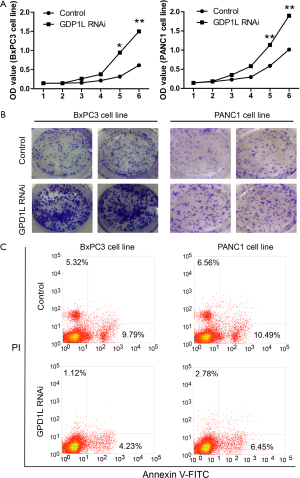
As GPD1L is an important prognostic factor in PDAC, we investigated the effects of GPD1L RNAi on proliferation, tumor formation and apoptosis. BxPC3 and PANC1 cells were transfected with GPD1L siRNA or control for 48 h. Then cell proliferation was measured by MTT assay. The results showed that GPD1L siRNA could promote cell proliferation (Figure 4A). Consistently, in the clone formation assay, GPD1L siRNA could increase clone numbers (Figure 4B). Moreover, in the Annexin V-FITC apoptosis assay, GPD1L siRNA could reduce apoptosis (Figure 4C). These results suggest that GPD1L reduction promotes aggressive tumor phenotypes and this is consistent with the fact that GPD1L down-regulation is associated with poorer prognosis.
Discussion
Recent advance in cancer biology has improved the treatment and prognosis for many types of cancer. However, the prognosis of PDAC remains very poor, suggesting its underlying pathogenesis mechanism is far from clear. Many studies support that microRNAs play a critical role in cancer. But it is common to see that differentially expressed microRNAs differ greatly among different studies and few of them could be cross-validated. It is likely that large parts of the microRNA signature may be noises without direct relevance. It is more likely for the cross-validated microRNAs to have true contribution in the pathogenesis of PDAC and therefore their target genes deserve further identification.
Here, we first compared the differentially expressed microRNAs in PDAC from three highly cited studies and identified 5 microRNAs which were reproducible across three different techniques. The relatively small overlap of microRNAs might be attributed to different methodology or high heterogeneity of PDAC (20). We further validated an up-regulation of 3-microRNA signature consisting miR155, miR181a and miR221 in PDAC. It’s important to note that miR155 and miR221 are also highlighted by a comprehensive meta-analysis of microRNA signature which uses two meta-review approaches (11). The good reproducibility of the 3-microRNA signature suggests that their target genes may play important roles in PDAC. Interesting, the miRDB prediction revealed that the 3-microRNA signature shared four target genes: CREBRF, GABRA1, GPD1L and REPS2. Among them, GPD1L was down-regulated in PDAC. In addition, the association between GPD1L down-regulation and poor prognosis could be cross-validated in three independent datasets. Thus, GPD1L is critical for the prognosis of PDAC.
Consistently, results from the BxPC3 and PANC1 pancreatic cancer cell lines suggest that the 3-microRNA signature inhibited GPD1L expression and GPD1L reduction promoted aggressive tumor phenotypes. Similarly, an independent study shows that microRNAs could coordinately inhibit tumor suppressor genes to promote pancreatic tumor growth and progression (13). Our findings are also consistent with a recent study showing that GPD1L mRNA level is reduced in head and neck squamous cell carcinoma and GPD1L down-regulation is associated with poor prognosis (21). In addition, previous study demonstrates that GPD1L could destabilize HIF1α by enhancing its hyper-hydroxylation and GPD1L expression is repressed under hypoxia (22). As hypoxia-induced HIF1α accumulation is a common feature in cancer including PDAC (23), GPD1L reduction might increase HIF1α accumulation to promote aggressive behaviors of PDAC. Thus, GPD1L is an important target gene for the 3-microRNA signature and GPD1L is a novel prognostic factor in PDAC.
In summary, we provide evidence showing that an up-regulated microRNA signature consisting of miR155, miR181a and miR221 could inhibit GPD1L expression to promote cell proliferation and clone formation, and reduce apoptosis in PDAC. Our study suggests that GPD1L is a novel prognostic factor and perhaps a therapeutic target in PDAC.
Acknowledgments
Funding: This work was supported by Zhejiang Provincial Natural Science Foundation of China (No. LY15H030016 and LY15H160056) and Wenzhou Science & Technology Bureau (No. Y20140696). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.22). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Boards of the first affiliated hospital of Wenzhou Medical University (Wenzhou, China) (No. 2016-099) and written informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:2140-1. [Crossref] [PubMed]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704-14. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007;297:1901-8. [Crossref] [PubMed]
- Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 2007;120:1046-54. [Crossref] [PubMed]
- Zhang Y, Li M, Wang H, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg 2009;33:698-709. [Crossref] [PubMed]
- Schultz NA, Dehlendorff C, Jensen BV, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014;311:392-404. [Crossref] [PubMed]
- Ma MZ, Kong X, Weng MZ, et al. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res 2013;32:71. [Crossref] [PubMed]
- Müller S, Raulefs S, Bruns P, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer 2015;14:94. [Crossref] [PubMed]
- Frampton AE, Castellano L, Colombo T, et al. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology 2014;146:268-77.e18. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Stratford JK, Bentrem DJ, Anderson JM, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med 2010;7:e1000307 [Crossref] [PubMed]
- Zhang G, Schetter A, He P, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One 2012;7:e31507 [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Chen DT, Davis-Yadley AH, Huang PY, et al. Prognostic Fifteen-Gene Signature for Early Stage Pancreatic Ductal Adenocarcinoma. PLoS One 2015;10:e0133562 [Crossref] [PubMed]
- Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer 2014;14:970. [Crossref] [PubMed]
- Yamano M, Fujii H, Takagaki T, et al. Genetic progression and divergence in pancreatic carcinoma. Am J Pathol 2000;156:2123-33. [Crossref] [PubMed]
- Feng Z, Li JN, Wang L, et al. The prognostic value of glycerol-3-phosphate dehydrogenase 1-like expression in head and neck squamous cell carcinoma. Histopathology 2014;64:348-55. [Crossref] [PubMed]
- Kelly TJ, Souza AL, Clish CB, et al. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol 2011;31:2696-706. [Crossref] [PubMed]
- Zhao X, Gao S, Ren H, et al. Hypoxia-inducible factor-1 promotes pancreatic ductal adenocarcinoma invasion and metastasis by activating transcription of the actin-bundling protein fascin. Cancer Res 2014;74:2455-64. [Crossref] [PubMed]


