Common SEP15 polymorphisms and susceptibility to cancer: a systematic review and meta-analysis
Introduction
Cancer is a global health problem that results in significant morbidity and mortality (1). Recent estimates indicate that approximately 4,292,000 new cancer cases and 2,814,000 cancer deaths are reported in China each year (2). To date, the exact mechanisms of carcinogenesis are poorly understood. An increasing number of studies have reported that cancer is a complex disease influenced by various environmental and genetic factors and their interactions (3,4). In addition, several genes have been associated with cancer susceptibility (5)
Selenoproteins are a class of proteins characterized by incorporation of selenium (Se) in the form of the amino acid (6). Studies investigating the association of Se with cancer susceptibility have described the role of Secis carcinogenesis. The 15 kDa selenoprotein (SEP15) and other members of the thioredoxin family (7) are a type of catalyzing agent that can regulate the cellular redox reaction and reduce cumulative oxidative stress, which has correlates with cell death and oncogenesis (8). A recent study has shown that SEP15 is unregulated in the prostate gland (9) and binds to uridine diphosphate (UDP)-glucose:glycoprotein glucosyltransferase (10-12), a regulatory protein of N-linked glycoprotein folding in the endoplasmic reticulum, which suggests that SEP15 plays an essential function in this particular pathway.
Recent studies have described the association between SEP15 polymorphisms and the risk of various cancers, including colorectal cancer (CRC) (13,14), lung cancer (LC) (15), breast cancer (BC) (16-18), and prostate cancer (PCa) (19-21). However, the results of these studies are conflicting and inconclusive, possibly due to clinical heterogeneity, different ethnic populations, and small sample sizes. To circumvent these limitations, we conducted a meta-analysis of the results of relevant studies to evaluate the association between SEP15 polymorphisms and cancer susceptibility.
Methods
Publication search eligibility of relevant studies
All case-control studies included in this study were queried from PubMed, EMBASE, Cochrane Library, and Web of Science using the following keywords: “SEP15 OR 15kDa selenoprotein” AND “variant OR mutation OR SNP OR polymorphism” AND “cancer OR tumor OR carcinoma OR malignancy OR neoplasms”. Only relevant studies in humans were included, and the language was restricted to English. In addition, references of eligible publications were searched manually. When certain data were not mentioned in the report, the corresponding author of the publication was contacted by e-mail. The most recent or complete articles with the largest number of subjects were selected from overlapping data of reports by the same authors. The last search was performed on December 17, 2016.
Eligible studies included in the meta-analysis met the following inclusion criteria: assessed the association between SEP15 polymorphisms (rs5859 and rs5845) and susceptibility to cancer; were case-control studies designed for human subjects; and provided useful data on genotype frequencies. Meanwhile, the exclusion criteria were as follows: duplicate data; clinical cases, comments, series, and reviews; and insufficient data. Studies published in languages other than English were also excluded. Articles with two or more case-control cohorts were regarded as two or more different studies.
Data extraction
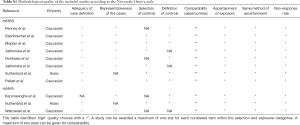
Two investigators independently reviewed the reports that fulfilled the selection criteria and extracted the following data: name of first author; year of publication; country of origin; ethnicity; and source of controls (population-based or hospital-based controls). The Newcastle-Ottawa scale (NOS) was applied to assess the quality of the studies included in our analysis. Different ethnic groups, including Caucasians, Asians, and Africans, were analyzed to assess the effects of SEP15 polymorphisms on cancer susceptibility.
Statistical analysis
The strength of the association between SEP15 polymorphisms and tumor susceptibility was assessed by the odds ratios (ORs) with 95% confidence intervals (CIs). For SEP15 polymorphisms, the susceptibility of dominant (MM + MW vs. WW), recessive (MM vs. MW + WW), co-dominant (MW vs. WW; MM vs. WW), and allele models (M vs. W) was evaluated, respectively (M: mutant allele; W: wild-type allele). Subgroup analyses were also conducted by ethnicity, source of control, and cancer type. The Hardy Weinberg equilibrium (HWE) was used to assess the genotype frequencies of SEP15 polymorphisms among the controls using the χ2 test. Meta-analysis was performed by the Mantel-Haenszel method in accordance with the Cochrane organization guidelines. The heterogeneity between datasets was evaluated by the heterogeneity index (I2) and the Cochran’s Q statistic (22). P-het <0.10 was considered as significant heterogeneity. The Fixed-effects model was applied when I2 was <50%, while random-effects model was used when I2 was >50% (23). Funnel plots were applied to test for publication bias (24). The forest plot was generated using the Review Manager software (RevMan, version 5.3; Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014). All analyses were performed with Stata software ver. 12.0.
Power analysis for association of SEP15 polymorphisms and tumor susceptibility was performed using the Genetic Power Calculator (13).
Results
Characteristics of studies
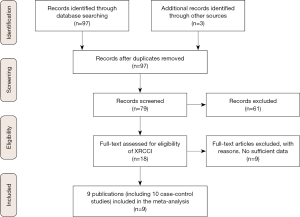
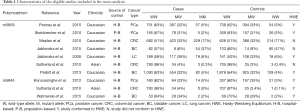
A total of 97 publications were identified after our initial search. After screening the titles and abstracts, 79 publications were excluded from the study, and 18 publications were selected for further full-text review. Nine studies were excluded because these were not case-control studies, did not describe SEP15 polymorphisms (rs5859 and rs5845) and cancer susceptibility, or did not provide detailed genotype data. We finally identified nine eligible publications, including 10 case-control studies (a total of 6,970 cases and 8,432 controls) that were subjected to our meta-analysis (Table 1) (14-21,25). The study selection processes are presented in Figure 1.
Full table
The publications included in this study were published from 2008 to 2016. For the SEP15 rs5859 polymorphism, seven studies comprising a total of 5,802 cases and 7,035 controls met the inclusion criteria. Six of these involved Caucasians and one included Asians. Six studies were population-based and one study was hospital-based. Additionally, two of the studies were performed on subjects with PCa, BC, and CRC, respectively, and one study was performed subjects with LC. There were three studies on the rs5859 polymorphism that did not conform to HWE (P<0.05) (14,17,21). For the SEP15 rs5845 polymorphism, three studies comprising a total of 1,168 cases and 1,397 controls were analyzed, two of which involved Caucasians, and one that included Asians; the controls in all three studies were hospital-based. In terms of cancer type, three studies were performed on PCa, BC, and CRC. In addition, we applied the NOS to evaluate the quality of these enrolled studies, which are presented in Table S1.
Results of the meta-analysis
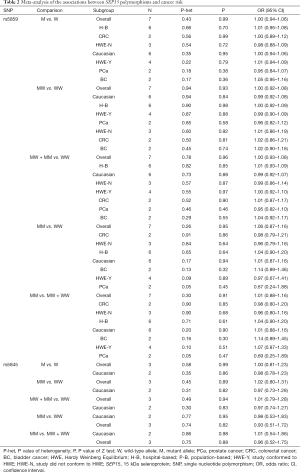
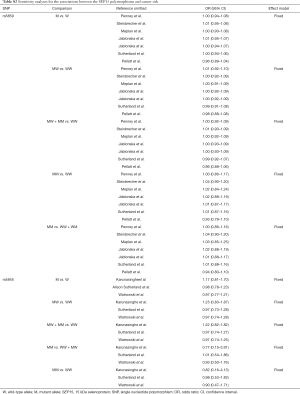
Overall meta-analysis of the studies on two SEP15 polymorphisms (rs5859 and rs5845) did not detect any significant association with cancer susceptibility (P>0.05; Table 2). No significant associations were observed in the stratification analyses by ethnicity, cancer type, HWE status, or source of control subjects (P>0.05; Table 2).
Full table
Test of heterogeneity, sensitivity analyses, and publication bias
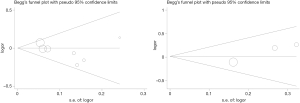
Overall comparison and subgroup analyses did not detect any significant heterogeneity among the studies included in the meta-analysis. We repeated our meta-analysis and omitted every study one by one to assess the effect of each eligible study on the results of our investigation. The pooled ORs for the effects of the rs5859 and rs5845 polymorphisms on cancer susceptibility indicated that our data were stable and reliable (Table S2). Begg’s funnel plot analysis indicated that our meta-analysis had significant symmetry and no publication bias (Figure 2), which was further validated by the Egger’s test (P>|t|=0.20 for the rs5859 polymorphism; P>|t|=0.12 for the rs5845 polymorphism).
Discussion
Se is a dietary micro-nutrient that is essential for human health (26,27), and cancer mortality has been inversely correlated with Se intake (27,28). The biological functions of Se are strongly associated with the aminoacid Sec, which is present in ~25 selenoproteins (29,30). Sec is incorporated into selenoproteins at the stem-loop structure of the 3’ untranslated region (3’ UTR), which requires an in-frame stop UGA codon and is recognized as a Sec insertion sequence (SECIS) by various trans-acting factors (29). These selenoproteins include the family of glutathione peroxidases (31): (I) selenoprotein P (SePP) that function as a transporter (32); (II) selenoprotein S, an endoplasmic reticulum protein involved in removing unfolded proteins; and (III) SEP15, another endoplasmic reticulum protein that also involved in the unfolded protein response (33,34). Not all identified selenoproteins have been characterized. However, several selenoproteins are known to possess redox functions and behave as antioxidants that reduce oxidative stress (29,31) Thus, it is possible that genetic variations in the form of polymorphisms in selenoproteins are associated with the risk for different types of cancer and/or oxidative stress. For example, regulatory elements within the 3’ UTR are essential for Se incorporation into selenoproteins and therefore, single nucleotide polymorphisms (SNPs) in gene regions corresponding to the 3’ UTR of selenoprotein mRNA shave the potential to influence selenoprotein expression. Indeed, minor allelic variants of rs5845 and rs5859 in SEP1 have functional consequences (35). Furthermore, polymorphisms have been associated with an increase in BC risk (35) and LC in smokers (15). Penney et al. (20) screened 1,286 cases and 1,267 controls and found that SEP15 polymorphisms were not significantly associated with PCa. Jablonska et al. (17) reported that patients with the SEP15 1125 AA had higher Se intake, whereas those harboring the GG or GA genotype, and a higher Se status were more susceptible to LC. Because the results of these studies are conflicting and inconclusive, we conducted the present meta-analysis. Overall, nine publications comprising 10 case-controls were enrolled, and the overall meta-analyses showed no significant association between SEP15 polymorphisms and cancer susceptibility (Table 2). When subgroup analyses were performed based on the source of the control subjects or cancer type, null results were also found (Table 2). Thus, our findings may serve as a foundation for the development of future investigations.
This study had a number of advantages. First, we have conducted a comprehensive literature search to identify eligible studies, thereby rendering our analysis as more persuasive and substantial. Second, the quality of the enrolled studies was assessed by NOS, and low-quality studies were generally excluded to raise the overall quality. Third, subgroup analysis was conducted according to cancer type, HWE status, and other specific study features for the purpose of further deepening our research for sources of data heterogeneity. Fourth, our results were adjusted according to the recognized formula, ensuring the accuracy of our results. In addition, the stability of these studies was further verified by sensitivity analysis, and publication bias was assessed by the Egger’s test and Begg’s funnel plot. This study also had several limitations that should be described. First, single case-control studies can only support a small test power and often provide false-positive, false-negative, or inconsistent conclusions, and the number of cases in the eligible studies were relatively small. Furthermore, some detailed information such as gender and histological type could not be obtained from these reports, and thus a more in-depth subgroup analysis could not be performed. Second, most of the eligible studies involved Caucasian patients, only a few studies included Asians, and none included Africans. Additional studies involving various populations are needed to obtain more convincing results. Third, most of the published studies were hospital-based and genotype distributions among the controls in some studies deviated from HWE.
In conclusion, our research shows that the SEP15 polymorphisms are not significantly associated with cancer susceptibility. Considering the limited studies in both overall and subgroup meta-analyses, larger sample sizes and higher quality studies are needed to validate these findings.
Full table
Full table
Acknowledgments
We thank Dr. Aihua Lin from the Public Health School of Sun Yat-sen University for technical guidance on statistical analysis.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Hoover RN. Cancer--nature, nurture, or both. N Engl J Med 2000;343:135-6. [Crossref] [PubMed]
- Pharoah PD, Dunning AM, Ponder BA, et al. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer 2004;4:850-60. [Crossref] [PubMed]
- Ponder BA. Cancer genetics. Nature 2001;411:336-41. [Crossref] [PubMed]
- Gresner P, Gromadzinska J, Jablonska E, et al. Expression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancer. Lung Cancer 2009;65:34-40. [Crossref] [PubMed]
- Rayman MP. Selenoproteins and human health: insights from epidemiological data. Biochim Biophys Acta 2009;1790:1533-40.
- Poerschke RL, Moos PJ. Thioredoxin reductase 1 knockdown enhances selenazolidine cytotoxicity in human lung cancer cells via mitochondrial dysfunction. Biochem Pharmacol 2011;81:211-21. [Crossref] [PubMed]
- Kumaraswamy E, Kim E. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J Biol Chem 2000;275:35540-7. [Crossref] [PubMed]
- Korotkov KV, Kumaraswamy E, Zhou Y, et al. Association between the 15-kDa selenoprotein and UDP-glucose: glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem 2001;276:15330-6. [Crossref] [PubMed]
- Labunskyy VM, Ferguson AD, Fomenko DE, et al. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose: glycoprotein glucosyltransferase. J Biol Chem 2005;280:37839-45. [Crossref] [PubMed]
- Labunskyy VM, Hatfield DL, Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life 2007;59:1-5. [Crossref] [PubMed]
- Méplan C, Hughes DJ, Pardini B, et al. Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis 2010;31:1074-9. [Crossref] [PubMed]
- Sutherland A, Kim DH, Relton C, et al. Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer. Genes Nutr 2010;5:215-23. [Crossref] [PubMed]
- Jablonska E, Gromadzinska J, Sobala W, et al. Lung cancer risk associated with selenium status is modified in smoking individuals by Sep15 polymorphism. Eur J Nutr 2008;47:47-54. [Crossref] [PubMed]
- Pellatt AJ, Wolff RK, John EM, et al. SEPP1 influences breast cancer risk among women with greater native american ancestry: the breast cancer health disparities study. PLoS One 2013;8:e80554 [Crossref] [PubMed]
- Jablonska E, Gromadzinska J, Peplonska B, et al. Lipid peroxidation and glutathione peroxidase activity relationship in breast cancer depends on functional polymorphism of GPX1. BMC Cancer 2015;15:657. [Crossref] [PubMed]
- Watrowski R, Dan CT, Fabjani G, et al. The 811 C/T polymorphism in the 3' untranslated region of the selenoprotein 15-kDa (Sep15) gene and breast cancer in Caucasian women. Tumour Biol 2016;37:1009-1015. [Crossref] [PubMed]
- Karunasinghe N, Han DY, Goudie M, et al. Prostate disease risk factors among a New Zealand cohort. J Nutrigenet Nutrigenomics 2012;5:339-51. [Crossref] [PubMed]
- Penney KL, Schumacher FR, Li H, et al. A large prospective study of SEP15 genetic variation, interaction with plasma selenium levels, and prostate cancer risk and survival. Cancer Prev Res (Phila) 2010;3:604-10. [Crossref] [PubMed]
- Steinbrecher A, Méplan C, Hesketh J, et al. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev 2010;19:2958-68. [Crossref] [PubMed]
- Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719-48. [PubMed]
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088-101. [Crossref] [PubMed]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003;19:149-50. [Crossref] [PubMed]
- Rayman MP. Selenium in cancer prevention: a review of the evidence and mechanism of action. Proc Nutr Soc 2005;64:527-42. [Crossref] [PubMed]
- Whanger PD. Selenium and its relationship to cancer: an update. Br J Nutr 2004;91:11-28. [Crossref] [PubMed]
- Combs GF, Gray WP. Chemopreventive agents: selenium. Pharmacol Ther 1998;79:179-92. [Crossref] [PubMed]
- Bellinger FP, Raman AV, Reeves MA, et al. Regulation and function of selenoproteins in human disease. Biochem J 2009;422:11-22. [Crossref] [PubMed]
- Lobanov AV, Hatfield DL, Gladyshev VN. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys Acta 2009;1790:1424-8.
- Brigelius-Flohe R. Glutathione peroxidases and redox-regulated transcription factors. Biol Chem 2006;387:1329-35. [Crossref] [PubMed]
- Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr 2005;25:215-35. [Crossref] [PubMed]
- Ferguson AD, Labunskyy VM, Fomenko DE, et al. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem 2006;281:3536-43. [Crossref] [PubMed]
- Labunskyy VM, Yoo MH, Hatfield DL, et al. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry 2009;48:8458-65. [Crossref] [PubMed]
- Hu YJ, Korotkov KV, Mehta R, et al. Distribution and functional consequences of nucleotide polymorphisms in the 3'-untranslated region of the human Sep15 gene. Cancer Res 2001;61:2307-10. [PubMed]