
HBx deletion-mutant combined with the reverse transcriptase enzyme/s* mutation is involved in HBV mediated liver cell proliferation and apoptosis
Introduction
One of the most common primary malignancies worldwide is Hepatocellular carcinoma (HCC). It ranks as the third most frequent cause of cancer-related deaths and it has been estimated that there are nearly 700,000 deaths per year (1). Chronic infection with the hepatitis B virus (HBV) is one of the major risk factors associated with HCC and it is estimated that infection with HBV results in more than 50% of all HCC cases (2,3). Because chronic hepatitis B (CHB) is highly prevalent in developing countries, there is a high incidence of HCC in poorer regions of the world (4). Host susceptibility, viral factors (including viral load, HBV genotype and viral mutations) and environmental factors are all known to contribute to HCC development and progression (3,5).
The HBV is a small, double-stranded circular 3.2-kb DNA virus. Its genome contains four overlapping open reading frames (ORFs) that encode for the envelope protein, the viral polymerase, X protein and the nucleocapsid (6). The HBV X gene encodes the HBx protein, which is a 154-amino acid protein that plays a crucial role in hepatocarcinogenesis. Many recent studies have investigated the effect of mutations in the HBV genome on HBV infection and associated outcomes. However, the different roles that HBx mutations play in hepatocarcinogenesis remain unclear. Previous studies revealed that the rtA181T mutation in HBV polymerase encodes a stop codon in the overlapping envelope proteins in the S region (sW172*). The occurrence of this mutation results in the truncation of the last 55 amino acids of the C-terminal hydrophobic region of the surface protein and this deletion is known to be associated with hepatocarcinogenesis (7,8). It has been suggested that intra-hepatic accumulation of mutant pre-S/S proteins caused by secretion defects could be the reason why such mutants are selected during antiviral therapy and why they are associated with HCC (9,10). The rtA181T/sW172* mutation has been associated with an increased risk of HCC in cirrhotic patients (11). Also, our previous studies showed that liver cancer tissues obtained from chronic HBV-infected human patients had an increased incidence of a particular HBx deletion mutation; i.e., HBx-d382 (which contains a deletion between 382–400 bp), which is potentially associated with the development of HCC (12). However, up until now studies have not been conducted to determine if HBx-d382 and rt/s* mutations co-exist in HCC patients. As part of this analysis, we aimed to investigate the co-existence of HBx-d382 and rt/s* mutations in HCC patients and the potential role of this combination of mutations in HCC.
In the present study, NA-resistant HCC samples were collected from CHB patients and sequenced to assess whether HBx-d382 and rt/s* co-existed in the recruited patients. We subsequently generated lentivirus constructs carrying different HBV mutations including rtA181T/sW172*, rtM204I/sW196*, HBx-d382, rtA181T/sW172*/HBx-d382 and rtM204I/sW196*/HBx-d382. These recombinant constructs were used to independently infect the human hepatocyte cell line, LO2. We monitored the effects of the mutations on apoptosis, cell proliferation, and the rate of colony formation in the transfected LO2 cells to better understand the influence of the HBV mutations in the development of HCC. Our data suggest that rt/s*/HBx-d382 mutants are more common in HBV-related HCC than in CHB and rt/s*/HBx-d382 triple mutants play a crucial role in HCC occurrence.
Methods
Study design and population
Between December 2015 and September 2016, 152 CHB and 33 NA-resistant HCC patients in Xiangya Hospital were enrolled in this study. The following characteristics were observed for patients included in this study: (I) all patients were HBsAg-positive and were receiving NAs for antiviral therapy (lamivudine, adefovir, entecavir, telbivudine, monotherapy, sequential or combined treatment); the course of treatment was greater than 1 year for each of the patients; (II) virological breakthrough and/or biochemical breakthrough occurred during antiviral treatment and HBV DNA was >104 copies/mL; (III) patients with HCV, HDV, HIV and other viral infections were excluded from the study; individuals with autoimmune, hereditary, metabolic liver disease were also excluded from the study.
DNA sequencing
To test for the presence of the RT/S*/HBx-d382 triple mutation in each of the patients enrolled in the study, we used DNA sequencing. DNA was extracted from patients with a HBV DNA titer >104 copies/mL and it was then used as a template to detect the presence of RT/S*/HBx-d382 via PCR product sequencing. The following HBV RT primers (covering the S region of the viral genome) were utilized to facilitate detection of HBV: A1: 5'-YCTCWSYCAYATCGTCAA-3', A2: 5'-GAGMCACAAAGGTTCCAC-3', B1: 5'-AGGCAGGATAGCCACATT-3' and B2: 5'-GCACCGAACATGGAGAAC-3'. Primers A1 and A2 were used in the first round of a nested PCR and primers B1 and B2 were used in the second round to test for the presence of the HBV RT region. The primer sequences used for HBx gene amplification were as follows: HBx-F: 5'-CCTACAGCCTCCTAAATCTCCTCCCCCAACTCCTCCC-3' and HBx-R: 5'-TTGGGGGAGGAGATTTAGGAGGCTGTAGGCATAAATTG-3'. Following PCR, a 5 µL PCR product from each sample was separated by 2% agarose gel electrophoresis. Positive PCR products were sequenced by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China) using an ABI 3730xl DNA Analyzer.
Cell culture
The normal human hepatocyte cell line LO2 (Chinese Academy of Science, Cell Biology of Shanghai Institute, Shanghai, China) was maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 100 µg/mL penicillin, 10% (v/v) fetal bovine serum (FBS), and 100 µg/mL streptomycin. The LO2 cells were incubated in a humidified 95% air and 5% CO2 incubator (MCO-15AC, SANYO, Japan) at 37 °C. The culture medium was changed daily and experiments were performed when the cells reached approximately 80% confluence.
Construction of plasmids
Construction of pcDNA3.1-1.3HBV plasmid
PCR primers were designed according to the whole HBV gene sequence (deposited in GenBank). The P1 and P2 primers were utilized to amplify the whole HBV gene and primers A1-D2 were utilized to amplify fragments A–D, respectively (Table 1). All primer sequences were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). HBV DNA, which was extracted from serum samples from the recruited patients, was used as a template to amplify the HBV gene (using P1 and P2 primers). The recombinant product was cloned into the pGEM-T vector to generate T-HBV recombinants. The resultant T-HBV recombinants were used as templates for subsequent PCR amplifications to amplify the A, B, C and D fragments (using the appropriate primers), respectively. The “A + B” and “C + D” gene fragments were generated by recombinant PCR. The resultant recombinant plasmid, pcDNA3.1-ABCD, was identified by PCR. The PCR product was digested with restriction enzymes and subsequently sequenced. The positive clones that were successfully screened were named pcDNA3.1-1.3HBV.

Full table
Construction of site-directed mutant plasmids
The primers used to amplify the requisite loci are listed in Table 2. Site-directed mutagenesis was performed using the pcDNA3.1-1.3HBV plasmid as a template. HBV mutants harboring rtA181T/sW172*, rtM204I/sW196*, HBx-d382, rtA181T/sW172*/HBx-d382 and rtM204I/sW196*/HBx-d382 mutations were constructed separately. An empty pcDNA3.1 vector was the control (pcDNA3.1-control).

Full table
Lentivirus packaging and cell infection
Lentiviral particles from HEK293T cells were co-transfected with virus packaging vectors as described previously (13). Briefly, HEK293T cells were seeded in a 10 cm dish. Using the calcium phosphate method, the cells were then triple-transfected with two packaging vectors (pCAG-VSV-G, a plasmid expressing the VSV-G envelope gene and pCMV R8.91, a plasmid expressing the gag/pol genes) and the lentiviral expression vector. The supernatant containing the viral particles was harvested at 48 h after transfection. It was then concentrated by ultracentrifugation (25,000 rpm, at 4 °C for 3 h). The lentivirus was subsequently added to the media to infect the LO2 cells.
Measurement of cell viability
LO2 cells were seeded at a density of 1×104 cells/well in a 96-well plate. The cells were infected with the different recombinant lentiviruses after 24 h, and then were divided into six groups: pcDNA3.1-control, pcDNA3.1-rtA181T/sW172*, pcDNA3.1-rtM204I/sW196*, pcDNA3.1-rtA181T/sW172*/HBx-d382, pcDNA3.1-HBx-d382, and pcDNA3.1-rtM204I/sW196*/HBx-d382. Infections were performed after 24, 48 and 72 h incubation periods. Fifty microliters of MTT solution (1 mg/mL) were added to each well. Then the plates were incubated at 37 °C for 4 h. The dye crystal was dissolved in 100 µL of DMSO following removal of the medium. To finish, optical density (OD) of each well at 490 nm using an ELISA micro-plate reader (BIO-RAD, USA) was immediately measured to determine cellular viability.
Cell apoptosis assay by flow cytometry
The LO2 cells were infected with lentivirus for 48 h using the following constructs: pcDNA3.1-control, pcDNA3.1-rtA181T/sW172*, pcDNA3.1-rtM204I/sW196*, pcDNA3.1-HBx-d382, pcDNA3.1-rtA181T/sW172*/HBx-d382, and pcDNA3.1-rtM204I/sW196*/HBx-d382. Next, the cells were collected and washed twice with cold PBS. Cellular apoptosis was detected by double-staining with Annexin V-FITC/7-ADD solution at room temperature. After 15 min, the stained cells were analyzed by flow cytometry (BD, San Jose, CA, USA). Apoptotic events were recorded as a combination of early apoptotic (Annexin V+/7-AAD−) and late apoptotic/dead (Annexin V+/7-AAD+) events. This experiment was conducted 3 times.
Colony formation assay
The cells were seeded evenly in 6-well plates and incubated for 24 h as a colony formation assay. Next, the cells were infected with lentivirus containing pcDNA3.1-control, pcDNA3.1-rtM204I/sW196*, pcDNA3.1-rtA181T/sW172*, pcDNA3.1-rtA181T/sW172*/HBx-d382, pcDNA3.1-HBx-d382, and pcDNA3.1-rtM204I/sW196*/HBx-d382 for 48 h. Following transfection, we changed the media and the cells were cultured for 14 days. The cells were fixed and then stained with Giemsa dye and methanol for 1 and 10 min, respectively. The number of colonies that were <1 mm was counted using a CX31 microscope (Olympus Corporation). The late clone formation efficiency was recorded as: (number of colonies/number of cells inoculated) ×100%. Each experiment was performed 3 times.
Western blot analysis
Whole LO2 cell lysates from cells infected for 48 h with lentivirus containing pcDNA3.1-control, pcDNA3.1-rtA181T/sW172*, pcDNA3.1-rtM204I/sW196*, pcDNA3.1-HBx-d382, pcDNA3.1-rtA181T/sW172*/HBx-d382 or pcDNA3.1-rtM204I/sW196*/HBx-d382 were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The DNA was then transferred to polyvinylidene difluoride (PVDF) membranes using the BioRad electrotransfer system. The membranes were immunoblotted with anti-p53, anti-p21 at 4 °C overnight, and anti-CyclinD1 antibodies. The resultant blots were subsequently incubated for 1 h with secondary anti-mouse or anti-rabbit antibody (Santa Cruz Biotechnology, 1:4,000) at room temperature. GAPDH (Millipore, 1:1,000) was used as a loading control.
Statistical analysis
Prism6 (GraphPad, SanDiego, CA, USA) was used to carry out statistical analyses. The results were expressed as the mean ± SEM. A one-way analysis of variance was used to determine the difference between the experimental group and control group. A P value <0.05 was considered as statistically significant.
Results
RT/ S*/HBx-d382 mutations in NA-resistant patients
To investigate whether RT/S* double mutations and HBx-d382 deletion mutations were present in the patients recruited during this study, samples from the 152 CHB and 33 NA-resistant HCC patients were sequenced for mutations. The results showed that 3 of the 152 CHB patients co-harbored an RT/S* double mutation and the HBx-d382 deletion mutation (2 cases of rtA181T/sW172*/HBx-d382, 1 case of rtM204I/sW196*/HBx-d382; the incidence of triple mutation was 1.97%). Of the 33 patients with HCC, 5 patients co-harbored an RT/S* double mutation and the HBx-d382 mutation (3 cases of rtA181T/sW172*/HBx-d382 and 2 cases of rtM204I/sW196*/HBx-d382; the incidence of triple mutation was 15.15%). The results showed that triple mutations can occur in patients. Also, the detection rate was relatively higher in patients with HCC compared to CHB (Table 3).
Full table
Construction of plasmids
The complete HBV ORF was amplified and used as a template for the synthesis of the “A + B” and “C + D” fragments (Figure 1A,B). A total of six clones were selected for PCR identification, and clones 3 # and 8 # were correctly identified by sequencing (Figure 1C). The “A + B” and “C + D” fragments were subsequently digested and ligated with similarly digested pcDNA3.1 (Figure 1D). A total of 24 clones were selected for PCR identification and clones 19 # and 22 # were subjected to restriction enzyme digestion with HindIII/NotI. Clone 22 # exhibited the requisite restriction enzyme digest pattern and was shown to contain the correct insert following sequencing (Figure 1E). We subsequently constructed the desired mutants by site-directed mutagenesis. PCR was performed with the appropriate primer pairs and the PCR products were digested with the same enzymes as those used to digest the recombinant pcDNA3.1-HBV plasmid. The sequence of the mutated plasmids was confirmed by sequencing.
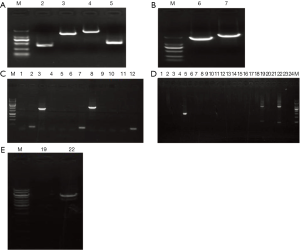
Fluorescence microscopy showed the effect of lentivirus infection in LO2 cells
Lentivirus LV5 is capable of expressing green fluorescent protein (GFP). This lentivirus strain was transfected into LO2 cells and the infection was observed by fluorescence microscopy. The results showed that green fluorescence was observed after transfection with different plasmids packaged by lentivirus, indicating that LO2 cells were successfully infected with lentivirus containing the different mutant plasmids (Figure 2).
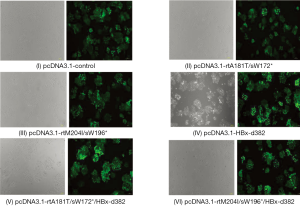
The HBx mutations enhance cellular proliferation in LO2 cells
We first constructed the following recombinant plasmids: pcDNA3.1-control, −rtA181T/sW172*, −rtM204I/sW196*, −HBx-d382, −rtA181T/sW172*/HBx-d382 and −rtM204I/sW196*/HBx-d382, to study the biological effects of HBx mutations in LO2 cells. We subsequently generated the recombinant lentiviruses (data not shown). Upon comparison with the pcDNA3.1-control, the HBx-d382, rtA181T/sW172*/HBx-d382 and rtM204I/sW196*/HBx-d382 mutations resulted in increased proliferation of LO2 cells (Figure 1). Interestingly, triple mutations (rtA181T/sW172*/HBx-d382 or rtM204I/sW196*/HBx-d382) resulted in accelerated LO2 cell growth compared with cells harboring double mutations (rtA181T/sW172* and rtM204I/sW196*) alone (Figure 3).
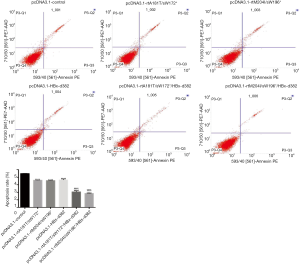
The HBx mutations repress cell apoptosis in LO2 cells
Flow cytometric analysis was employed to determine whether the HBx mutations repressed apoptosis of LO2 cells. As indicated in Figure 2, apoptosis analysis revealed that the HBx mutations (rtA181T/sW172*, rtM204I/sW196*, HBx-d382, rtA181T/sW172*/HBx-d382 and rtM204I/sW196*/HBx-d382) significantly decreased the rate of apoptosis. Furthermore, the combination of rtA181T/sW172* or rtM204I/sW196* with HBx-d382 resulted in a reduced rate of apoptosis compared with rtA181T/sW172* or rtM204I/sW196* alone (Figure 4).
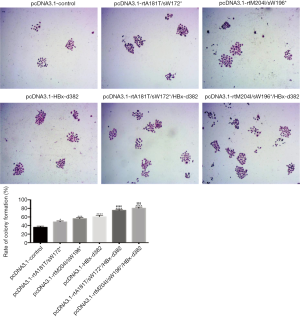
HBx mutations promote colony formation of LO2 cells in vitro
A colony formation assay was performed following infection of LO2 cells with LV-packaged recombinant plasmids carrying rtA181T/sW172*, rtM204I/sW196*, HBx-d382, rtA181T/sW172*/HBx-d382 or rtM204I/sW196*/HBx-d382 mutations. The results demonstrated that HBx mutations significantly promote colony formation of LO2 cells (Figure 5) and the triple mutant LO2 cells (rtA181T/sW172*/HBx-d382 or rtM204I/sW196*/HBx-d382) exhibited enhanced colony formation compared with cells harboring the rtA181T/sW172* or rtM204I/sW196* mutations alone (Figure 5).
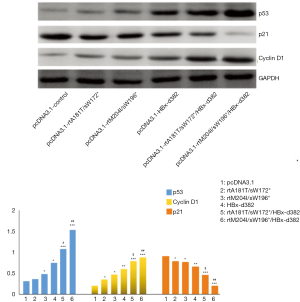
HBx mutations modulate the expression of p53, p21, cyclin D1, myc and NF-κB in LO2 cells
Western bolt analysis was utilized to analyze the expression of p53, p21, cyclin D1, myc and NF-κB to examine the effect of HBx mutations on LO2 cells. This analysis revealed an increase in p53 and cyclin D1 levels and a reduction in p21 levels in LO2 cells harboring the HBx mutations (Figure 6). Importantly, when the HBx-d382 mutation was combined with rtA181T/sW172* or rtM204I/sW196* there was a significant enhancement in expression levels compared with rtA181T/sW172* or rtM204I/sW196* mutations alone (Figure 6). These results suggest that the promotion of LO2 cell proliferation caused by HBx mutation can be attributed to the repression of apoptosis.
Discussion
The HBx protein is a tumorigenic protein that has been associated with HCC progression. HBV mutations are closely related to HCC development and progression (14,15). The detailed mechanisms underpinning the role of HBx mutations in hepatocarcinogenesis, however, remain unclear. In this study, we observed that mutants containing HBx-d382 combined with rtA181T/sW172* or rtM204I/sW196* promoted hepatoma cell proliferation markedly and repressed apoptosis in LO2 cells. The HBx protein is known to decrease cell proliferation by inducing both apoptosis and late G1 arrest in a p53-dependent and -independent manner (16). We also observed that the co-occurrence of the HBx-d382 mutation with rtA181T/sW172* or rtM204I/sW196* enhanced p53 and cyclin D1 expression and downregulated the expression of p21. We concluded that the occurrence of the triple afore-mentioned mutations promote hepatoma cell tumorigenicity by activating the p53-dependent signaling pathway.
The HBV CP region overlaps with the coding sequence for the C-terminus in the HBx gene (17). The HBx protein is an important oncoprotein in HCC pathogenesis (18) and mutations in the C-terminal coding sequence are likely to alter the biological function of this protein, thereby resulting in HIV-related HCC. HBV core promoter mutations have been shown to be related to severe hepatic inflammation and liver cirrhosis (19). The significance of the rtA181T/sW172* mutation in the clinical situation may be greater than the associated sensitivity to antiviral drugs (20,21). The latter phenomenon is caused by secretory defects and a dominant negative effect in relation to wild-type HBV virion secretion (9). In addition, the rtA181T/sW172* mutant exerts cross-resistance to both lamivudine and adefovir (22) and the prevalence of HBx mutations was significantly higher in HCC patients compared to chronic hepatitis patients (23).
The HBx gene consists of two functional domains: a C—terminal co-activation or transactivation domain and an N—terminal negative regulatory domain (24). Currently, the mechanisms underlying the initiation of hepatocarcinogenesis by HBx are not completely understood. A previous study showed that HBx mutations that result in a COOH—terminal truncation occur more frequently in HCC than in non-tumor tissues (25). A COOH-terminal deletion mutation in HBx results in inhibitory effects in relation to cell proliferation and transformation and reduced transcriptional activity. It has been suggested that this deletion mutation results in the activation of the myc oncogenes, thereby promoting tumor migration and proliferation (26). Furthermore, HBx-d382 is capable of promoting proliferation and anchor-independent growth of LO2 cells as well as enhancement of Cyclin D1 gene expression (27). In our study, we found that the HBx-d382 combined with rtA181T/sW172* or rtM204I/sW196* occurs in some NA-resistant patients. The incidence of this co-occurrence was high, especially in patients with HCC. Furthermore, in vitro experiments revealed that triple mutants displayed upregulation in the expression of p53 and cyclin D. However, the mechanisms that underpin the activation of p53 signaling by HBx mutants remain unclear. Further studies are required to elucidate the mechanisms underlying the activation of p53 signaling in the HBx-d382/rtA181T/sW172* and HBx-d382/ rtM204I/sW196* triple mutants.
In summary, we discovered that the HBx-d382 mutant combined with rtA181T/sW172* or rtM204I/sW196* markedly promoted hepatoma cell proliferation and repressed apoptosis in LO2 cells. Furthermore, we also observed that HBx-d382 combined with rtA181T/sW172* or rtM204I/sW196* resulted in a promotion in the expression of p53 and cyclin D1 and an inhibition in the expression of p21 compared with HBx-d382 only. In summary, this study showed that HBx mutations have an important role in the progression and aggressiveness of HCC.
Acknowledgments
Funding: This study was supported by Hunan Natural Science Foundation (2017JJ3510), which is the basic research program of Science and Technology Department of Hunan Province.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.43). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-2. [Crossref] [PubMed]
- Bréchot C, Gozuacik D, Murakami Y, et al. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol 2000;10:211-31. [Crossref] [PubMed]
- Chen CJ, Yang HI, Su J, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 2006;295:65-73. [Crossref] [PubMed]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264-73.e1. [Crossref] [PubMed]
- Liaw YF, Chu CM. Hepatitis B virus infection. Lancet 2009;373:582-92. [Crossref] [PubMed]
- Locarnini S. Molecular virology of hepatitis B virus. Semin Liver Dis 2004;24:3-10. [Crossref] [PubMed]
- Lai MW, Yeh CT. The oncogenic potential of hepatitis B virus rtA181T/ surface truncation mutant. Antivir Ther 2008;13:875-9. [PubMed]
- Lai MW, Huang SF, Hsu CW, et al. Identification of nonsense mutations in hepatitis B virus S gene in patients with hepatocellular carcinoma developed after lamivudine therapy. Antivir Ther 2009;14:249-61. [PubMed]
- Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology 2008;48:88-98. [Crossref] [PubMed]
- Huang CH, Yuan Q, Chen PJ, et al. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J Hepatol 2012;57:720-9. [Crossref] [PubMed]
- Yeh CT, Chen T, Hsu CW, et al. Emergence of the rtA181T/sW172* mutant increased the risk of hepatoma occurrence in patients with lamivudine-resistant chronic hepatitis B. BMC Cancer 2011;11:398. [Crossref] [PubMed]
- Zhu P, Tan D, Peng Z, et al. Polymorphism analyses of hepatitis B virus X gene in hepatocellular carcinoma patients from southern China. Acta Biochim Biophys Sin (Shanghai) 2007;39:265-72. [Crossref] [PubMed]
- Zhao A, Zeng Q, Xie X, et al. MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J Genet Genomics 2012;39:29-35. [Crossref] [PubMed]
- Guo X, Jin Y, Qian G, et al. Sequential accumulation of the mutations in core promoter of hepatitis B virus is associated with the development of hepatocellular carcinoma in Qidong, China. J Hepatol 2008;49:718-25. [Crossref] [PubMed]
- Yeh CT, So M, Ng J, et al. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology 2010;52:1922-33. [Crossref] [PubMed]
- Sirma H, Giannini C, Poussin K, et al. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene 1999;18:4848-59. [Crossref] [PubMed]
- Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res 2007;127:164-76. [Crossref] [PubMed]
- Pang R, Lee TK, Poon RT, et al. Pin1 interacts with a specific serine-proline motif of hepatitis B virus X-protein to enhance hepatocarcinogenesis. Gastroenterology 2007;132:1088-103. [Crossref] [PubMed]
- Chen CH, Hung CH, Lee CM, et al. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology 2007;133:1466-74. [Crossref] [PubMed]
- Qi X, Xiong S, Yang H, et al. In vitro susceptibility of adefovir-associated hepatitis B virus polymerase mutations to other antiviral agents. Antivir Ther 2007;12:355-62. [PubMed]
- Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol 2006;44:422-31. [Crossref] [PubMed]
- Lee YS, Suh DJ, Lim YS, et al. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology 2006;43:1385-91. [Crossref] [PubMed]
- Li W, Chen G, Yu X, et al. Accumulation of the mutations in basal core promoter of hepatitis B virus subgenotype C1 increase the risk of hepatocellular carcinoma in Southern China. Int J Clin Exp Pathol 2013;6:1076-85. [PubMed]
- Gong DY, Chen EQ, Huang FJ, et al. Role and functional domain of hepatitis B virus X protein in regulating HBV transcription and replication in vitro and in vivo. Viruses 2013;5:1261-71. [Crossref] [PubMed]
- Wang D, Cai H, Yu WB, et al. Identification of hepatitis B virus X gene variants between hepatocellular carcinoma tissues and pericarcinoma liver tissues in Eastern China. Int J Clin Exp Pathol 2014;7:5988-96. [PubMed]
- Tu H, Bonura C, Giannini C, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res 2001;61:7803-10. [PubMed]
- Fu X, Tan D, Hou Z, et al. miR-338-3p is down-regulated by hepatitis B virus X and inhibits cell proliferation by targeting the 3'-UTR region of CyclinD1. Int J Mol Sci 2012;13:8514-39. [Crossref] [PubMed]


