
Canonical and selective approaches in exosome purification and their implications for diagnostic accuracy
Introduction
Cell to cell signalling is crucial for maintaining homeostasis in multicellular organisms. Information exchange can be accomplished through direct cell-to-cell contact or through transfer of different secreted molecules. In addition, most eukaryotic cells release membrane derived vesicles that can impact both neighbouring and distant cells (1). Extracellular vesicles (EVs) are classified based on their cellular origin and/or biological function or based on their biogenesis. As determined by their biogenesis, the three main classes of EVs are exosomes, microvesicles and apoptotic bodies (2) (Figure 1). Exosomes are cell-derived vesicles present in many body fluids (blood, urine, milk, ascites and cerebrospinal fluid) ranging in size from 30 to 150 nm (3). First reports dealing with the formation of EVs emerged in the 1980s from two independent research groups. They described the formation of multivesicular bodies in reticulocytes that released such vesicles into the extracellular space (4,5). One decade later, the release of these vesicles has been described for lymphocytes and dendritic cells through a similar route (6,7). Since then, exosomes have been purified from nearly all mammalian cells including primary cells of the immune (8) and nervous system (9,10) and stem cells (11,12). Interestingly, this mechanism is not exclusively restricted to mammals, but secretion of EVs has been identified in plants (13), lower eukaryotes (14,15) and prokaryotes (16).
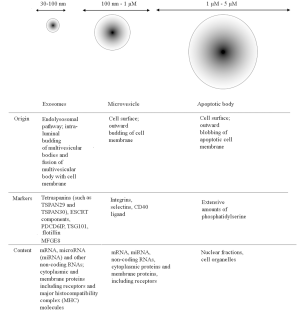
Numerous studies deal with physiological and pathological roles of EVs and exosomes in living organisms. So far they have been linked with transfer of proteins, lipids, nucleic acids and possibly active metabolites (1,17-20), cell self-renewal (11), tissue repair (21), immune surveillance (6,22) and blood coagulation (23). Besides these physiological processes, the role of exosomes has been shown in genesis of tumors (24,25) and their metastasis (26), spread of pathogens (27,28), transfer of toxic peptides linked with neurodegenerative diseases such as amyloid-β-derived peptides (29) and α-synuclein (30), as well as the spread of the abnormal pathogenic cell surface prion protein PrPC (31). EVs can also mediate inter-species horizontal transfer of coding mRNAs and the consequent translation of human genetic material into murine proteins (32), resulting in in vivo protective effect against cisplatin-induced kidney injuries (33) and liver morphological and functional recovery (34). Exosome exert their effect in a pleiotropic manner by directly activating cell surface receptors for proteins and merging with bioactive lipids of the receiving cell, a process which results in direct delivery of their internal cargo in the host. This can include transcription factors, oncogenes, small and large non-coding regulatory RNAs [such as microRNAs (miRNAs)], mRNAs, and infectious particles (1,11,25,32,35). Due to their involvement in a myriad of cellular processes, exosomes could be considered as multifunctional signalling complexes for controlling fundamental cellular and biological processes.
Several proteomics studies performed on different types of material (exosomes from primary cells, cell cultures, tissue cultures and biological fluids) yielded extensive data on protein abundance on different types of vesicles and these data are available in several public on-line repositories such as Vesiclepedia (www.microvesicles.org) (36), EVpedia (www.evpedia.info) (37) and ExoCarta (www.exocarta.org) (38). Protein content of EVs is made up of pan-exosome markers common for most of exosomes, but also proteins and protein post-translational modifications that specifically reflect the cellular origin and the mechanism of secretion (39-41). In general, exosomes contain cytoskeleton, cytosolic, heat-shock and membrane proteins, as well as proteins involved in vesicle trafficking and protein-lipid complexes (42). However, the proteomic profiles obtained in different studies depended much on the manner of exosome isolation and, since different methods yielded different EV populations and exosome sub-populations, the proteomic data are hard to compare.
Due to their involvement in numerous physiological and pathological events, cell derived exosomes in bodily fluids represent a unique source of clinically relevant and non-invasive biomarkers. The biomolecule content present in exosomes mirrors that of the parent cells and, therefore, exosome analysis may provide valuable information about presence of aberrant processes in the cells from which they are originated. Particularly peripheral blood and urine are rich and easily accessible sources of circulating exosomes. Analysis of exosomal content in peripheral blood provided access to exceptional amounts of biomarkers of diagnostic and prognostic value and, consequently, exosomes seem a particularly convenient target for liquid biopsy procedures (43). However, present exosome isolation approaches based on ultracentrifugation (UC), UF, and precipitation, are often inefficient, hard to standardize, thereby creating variability in sample quality and often leading to loss of important biomolecules (44).
State of the art
Much of our understanding of exosome biology has been obtained from heterogeneous or impure exosome preparations. Many of the published studies fail to assess the purity of the isolated exosome populations before performing functional assays or omics analyses despite the precise indications listed by the scientific community (36,45,46). This condition jeopardizes the interpretation of findings and the possibility to use EVs for therapeutic applications (47). The priority is therefore the identification of more reliable methodologies for the preparation of homogeneous exosome populations not contaminated with further membranous particles—such as apoptotic bodies (400–2,500 nm), shed microvesicles (50–1,500 nm), oncosomes (>1,000 nm)—released by cells into body fluids and sometimes sharing some of the “standard exosome” biomarkers (48-51).
Commonly purification methods include UC, density gradient (DG) centrifugation, chromatography, filtration, polymer-based precipitation and immunoaffinity (IA) (50). Both the quantity and the quality of the purified EVs critically depend on the applied purification strategies (50) which can yield exosomal preparations contaminated with vesicles and protein complexes, exosome with damaged membrane integrity, or the simple loss of exosomal fractions during the purification process. All these conditions can result in misleading data interpretation of the genomics and proteomics profiles.
Despite the several commercial polymer-based kits available on the market and that promise the fast precipitation of the exosome fraction, the gold standard for exosome isolation remains based on differential UC (52), mostly integrated with DGs, UF, size exclusion chromatography (SEC) or IA steps (50). UC exploits multiple centrifugation steps performed at increasing centrifugal strength to sequentially pellet cells (250–400 ×g), cell debris (2,000 ×g), microvesicles (10,000–20,000 ×g), and exosomes (100,000–150,000 ×g). Of course, the characteristics of the original samples strongly influence the purification final output. Cell culture supernatants represent a relatively simple and homogeneous starting material in comparison to body fluids characterized by high-abundance of contaminating proteins, lipoprotein particles and EVs originated by different cell types as well as by physical properties that might interfere with standard isolation protocols. Because this complexity represents a further element that can perturb EV data comparison, also the standardization of the protocols for the preparation of biological samples to undergo EV fractionation is under discussion (51,53). In parallel, the community search for specific biomarkers that would enable the selective and straightforward IA purification of exosome populations directly from biological samples (54) (Table 1).

Full table
UC and DG UC
UC has been for long time considered the method of choice when it comes to exosome isolation. In UC, centrifugal force is applied to a mixture of macromolecules in solution sedimenting them according to density. This involves applying to the sample a high g-force (100,000 ×g or greater) to pellet the vesicles. The most widely accepted method for exosome isolation is differential centrifugation (55,56). Successive centrifugation steps with increasing speeds of centrifugal force are intended to pellet consecutively the apoptotic bodies and cell debris, the shedding vesicles and the exosomes. Since UC was considered the “gold standard” in exosomal purification, it still accounts for the most commonly exosome isolation technique employed by users in exosome research (54). Although this methodology should be relatively straightforward, quantity and quality of the obtained material are highly sensitive to several parameters such as g-force, rotor type (fixed angle or swinging bucket), the angle of rotor sedimentation, the radius of centrifugal force, the pelleting efficiency (rotor and tube k-factors), and the solution viscosity (44,57,58). Viscosity difference among various biological fluids used for exosome purification influences the internal friction within the solution resulting in variable optimal centrifugation velocities needed for pelleting (50,59). The design of the centrifuge rotor also influences the efficacy of pelleting, as swing bucket rotors show lower pelleting efficacy but appear to produce better resolution of vesicles with similar sedimentation coefficients (57). Beside rotor design, ultracentrifuge clearing factor (k-factor) plays an important role. K-factor could be used to predict the time required for pelleting as it results from the correlation between k-factor and the sedimentation coefficient (in Svedberg units):. Many applied protocols use the same pelleting time and speed, but the difference in k-factor of the used devices leads to drastically different yields of exosomes from the same samples (48). It is therefore essential to adjust the centrifugation time to compensate for different rotor types. All these factors are hard to standardize and control and often account for the observed dissimilarity in data even when same protocols are applied. Numerous studies have indicated that UC can either result in the incomplete sedimentation of vesicles or the sedimentation of non-vesicular materials (48). On the top of that, it is still unclear how prolonged pelleting against solid surface affects the fluid membrane in terms of integrity and vesicle content, although the functionality of gradient-purified exosomes has been thoroughly described in some cases (60).
Improvements of classical UC, such as DG UC are now available. In DG UC, exosome separation depends on their size, mass, and density. A reduced volume of the sample is first loaded on a DG medium formed in a centrifuge tube in which density decreases progressively from the bottom to the top, then the DG is subjected to an extended round of UC. The most common DG-based methods exploit either sucrose or iodixanol [OptiPrepTM (Sigma-Aldrich, St. Louis, MO, USA)] (61). In particular, iodixanol-based gradients have been described improving the separation of exosomes from viruses and small apoptotic bodies (62). Upon applying the centrifugal force, solutes move through the DG medium towards the bottom according to their respective specific sedimentation rates, generating discrete solute zones. The separated exosomes can then be conveniently recovered by differential fraction collection. Both continuous and discontinuous gradients are used (63). The limiting factor of DG is the restricted volume that can be loaded, a factor which in turns limits the preparative capacity of the method (64). Furthermore, this protocol requires long running time to reach equilibrium, with reports spanning from 16 to 62–90 h (65,66). DG can lead to incomplete sedimentation of all exosomal fractions or artefacts from contaminating material co-migrated in the same density fractions due to inadequate sedimentation time. Furthermore, since the separation is achieved based on density, exosomal fraction can contain other vesicles of different origin but similar characteristics, such as cholesterol vesicles (LDL/HDL) (50,67-69).
Size-based isolation techniques
Different approaches of vesicle separation according to their size have been employed including UF, SEC, ion-chromatography, and filed-flow fractionation (FFF) (64).
UF in exosome preparation is no different form standard applications of this technique in other analytical fields. The principle of UF relies on the possibility to separate the particles present in a complex solution by exploiting their dimension. Based on their diameter, exosomes can be separated using membrane filters of defined size-exclusion limits (70) usually in a multi-step process designed to specifically remove contaminants with different characteristics. UF requires less time than UC and no special equipment is necessary for performing the separation (71). However, the exerted pressure may cause deformation and breakage of large-size vesicles influencing the results of down-stream analysis (72). Recently, it has been demonstrated that the filter material can significantly influence the final yields, with 10 kDa regenerated cellulose filters being the most efficient solution for exosome purification from plasma and urine (63).
Nano-membrane concentrators have been successfully employed for purification of exosomes from urine, more rapidly and effectively as UC (73). Short centrifugation time enabled the isolation of intact vesicles from as little as 0.5 mL of urine, as confirmed by electron microscopy and Western blot. This approach simplified and accelerated the isolation of urinary exosomes from clinical samples demonstrating its potential diagnostic applicability.
Sequential filtration for exosome isolation has been used mainly for cell culture supernatant as source of exosomes. This approach implies passing the source of exosomes through a series of membranes with different molecular weight cut-off to eliminate cells and cell debris (100 nM) and then soluble proteins (500 kDa) while in the final step the exosome containing solution is concentrated using a 100 kDa membrane (64). Sequential filtration allows the isolation of exosomes with high purity and apparent functional integrity as the result of low manipulation forces and in combination with sucrose DG has been successfully employed for purification of therapeutic exosomes for clinical trials (74,75).
SEC has always been a popular means for EV enrichment because it separates efficiently them from the bulk of soluble macromolecules present in biological samples using mild physical conditions which let unaffected vesicle integrity and structure (50). In SEC, porous stationary phase is utilized to sort macromolecules and particulate matters out, according to their size. Components in a sample with small hydrodynamic radii are able to pass through the pores, thus resulting in late elution. Components with large hydrodynamic radii including exosomes, are excluded from entering the pores. SEC was used for isolation of intact exosome from mesenchymal stem cells conditioned medium. Structural integrity of exosomes isolated in such a way was confirmed by transmission electron microscopy while other features such as size distribution and molecular markers were confirmed by dynamic light scattering (DLS), nanoparticle tracking analysis (NTA) and Western blot respectively (76). SEC is also useful to remove OptiPrep remnants from gradient-purified exosomes (63).
The net EV ζ-potential was exploited to induce protamine-dependent EV precipitation from serum, saliva, and cell culture media without the need for UC (77). This bulk separation based on total charge should not undermine the fact that the proteasome displayed on exosomes would correspond probably to unique fingerprints for each vesicle sub-class and, consequently, would have very peculiar surface chemical characteristics, such as the net charge. However, there are only few reports of conventional ion-exchange chromatographic approaches applied to exosomes. Recently, Kim et al. (78) demonstrated that anion exchange chromatography—in contrast to cation-based resins—is suitable for binding EVs and convenient for material concentration and consequently for scaling-up the sample volumes used in preparative purifications. However, the authors did not evaluate if non-vesicle contaminants co-eluted in the unique EV-containing peak recovered at 0.5 M NaCl. The results of the limited characterization of the EV surface biomarkers suggested that vesicles with different characteristics co-existed even though the complexity of the original sample (cell culture supernatant) was reduced. In a preliminary work (79), anion-exchange monolith resin seemed to provide better resolution and distinct EV elution peaks. Unluckily, also in this case no sufficient biochemical characterization of the peak contents was performed to assess the quality of the purified fraction. In a further work in which monolith anion exchange chromatography was used, it appeared that EVs can co-elute with a fraction corresponding to virus particles (80). Altogether, these reports indicate that ion exchange can be useful for a preliminary enrichment of EVs from large volumes of samples but are insufficient to infer conclusions about fraction purity and possibility to separate among EV subclasses.
FFF allows the field-dependent separation of the particles present in a fluid according to their variable mobility under the exerted force conditions. FFF separation is made according to particle hydrodynamic diameters and can discriminate very precisely among complex sample components over a wide colloidal size range. Asymmetrical flow field-flow fractionation (AF4) technique was proposed originally 10 years ago to isolate macrovesicles (81,82). Low resolution AF4 was effective in mild purification of miRNA-containing fractions (exosomes and high-density lipoproteins) directly from patient sera (83). However, only recently its potential for purification and quantification of different exosome sub-populations was confirmed by means of studies which coupled it to multi-detection systems based on UV, dynamic and multi angle light scattering, and transmission electron micrographs (84-87). The apparent limit of this technology, although very efficient in terms of sub-group separation, is that so far proved to be suitable only for analytical characterization of small samples pre-purified using a conventional protocol and not as an actual preparative method. Furthermore, it relies on the availability of very expensive pieces of equipment and highly trained personnel necessary to analyse the data sets.
Precipitation of exosomes
Settling out of exosomes from biological fluids can be achieved by altering their solubility or dispersity. This is usually achieved by water-excluding polymers such as polyethylene glycol (PEG) (71). Water-excluding polymers retain the water and force less-soluble components out of the solution (71). Samples containing EVs are incubated, usually at low temperature (4 °C) and for relatively long time (overnight), with a precipitation solution containing polymers such as PEG 8000, after which the exosome-enriched precipitate is isolated by means of either low-speed centrifugation or filtration (71). Precipitation of exosomes is easy to do and it does not require any kind of specialized equipment, only that cells and cell debris are removed from samples. This simplicity enables straightforward integration of the method in both research labs and clinical applications (72). Currently, several commercial exosome precipitation kits compatible with different biological fluids, such as plasma, serum, ascites, urine, cerebrospinal fluid, and culture medium are available on the market (88). These systems have been largely employed for the purification of exosomes, for instance from urine samples yielding high quantities of miRNAs and mRNAs (89). The major downside of polymer based exosome precipitation is the abundant co-precipitation of non-exosome contaminants, such as proteins and polymeric materials (54). This shortcoming has been repeatedly evidenced by comparative assessments of exosome purification protocols (see below).
Affinity and IA purification
As it has been pointed out, UC, DG, SEC and polymer-based precipitation do not preferentially isolate specific EV sub-classes (50). It means that by using conventional purification methods potentially informative exosomes, such as the tumor-derived ones that never exceed 10% of the total circulating exosomes (50), will represent a marginal fraction even in a highly pure fraction. The consequence is that significant variations of cargo biomarkers could remain unappreciated because of the dilution effect of the “informative vesicles” in the bulk of physiological EVs and this condition is clearly exasperated when the sample is highly contaminated by other biological components. Affinity chromatography can increase recovery specificity if the bound molecules are biomarkers expressed solely on the outer surface of (sub-groups of) the target vesicles. For instance, lectins and heparin can bind EVs (90-93) and several lectins have been identified as having high affinity towards saccharide residues on the surface of urinary exosomes and have been used successfully as means for exosome precipitation (85). Downstream analyses such as atomic force microscopy, DLS, Western blot and RNA analysis confirmed physical and molecular properties of isolated vesicles (85) but lectins have not been exploited to selectively enrich some exosome sub-classes. Nevertheless, the recent demonstration that fibronectin exposed on myeloma cell exosomes is the molecule that acts as the ligand for heparan sulfate (94) opens the opportunity to more selective purification methods.
Binders specific for membrane lipids have been used for separating exosome sub-populations according to their membrane composition. The phosphatidylserine-binding Tim4, a protein expressed on macrophages, was immobilized on magnetic beads for enabling IA capture. The approach was successfully used but only to capture total EV fractions, from both conditioned media and biofluids (95). A more interesting application has been reported by Tan et al. [2014]. The authors showed that cholera toxin B chain (CTB) and annexin V (AV), which bind to GM1 ganglioside and phosphatidylserine, respectively, succeeded in isolating two distinct EV groups from plasma. The approach has been further developed by the addition of Shiga toxin B chain (STB)—specific for globotriaosylceramide—as a new EV-binding substrate and sequential immunopurification resulted in the isolation of EV populations with unique contents (96,97). The approach has been recently used to isolate three different exosome populations starting from patient ovarian ascites (97).
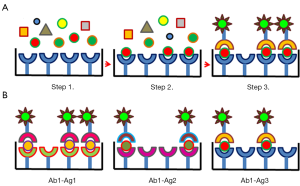
IA purification exploits the highly selective and strong interaction between antibodies and ligands displayed on the vesicle surface. Due to the extremely specialized structure that antibodies have developed for recognizing specifically their antigens, IA capture possesses the potential capacity to differentiate between minimal conformational diversities. However, the implementation of immunopurification as the standard method for the selective isolation of exosome sub-populations (for instance derived from cells belonging to different tissues or differing for maturation stage) will be possible only when reliable biomarkers for exosome subclasses will be identified and the corresponding antibodies will be produced and validated. In a seminal work, Clayton et al. (98) succeeded in demonstrating that exosomes derived by different antigen-presenting cells displayed specific combinations of biomarkers on their surface and that immunocapture enabled to recover analytical amounts of vesicles on the surface of magnetic beads. These biomarkers were often shared but varied in terms of expression level among vesicles originated by different cell types. Apart from well characterized antigens [the members of the human epidermal growth factor receptor family (99), EpCAM, Mart-1, TYRP2] displayed on the outer side of exosomes generated by different tumors, even at the present IA exploits mostly either antibodies against exosome universal biomarkers such as CD63 for the terminal purification step (94,100-102) or is limited to capture EVs originated from some well-characterized cell types which possess specific rather than exclusive molecular markers. For instance, anti-EpCAM antibodies have been successfully used to discriminate epithelial tumor-specific from other circulating exosomes assuming that healthy cells do not express such a biomarker (44). In another application, MHC class II was targeted to immunoprecipitate exclusively the exosomes derived from B cells from a pre-purified EV pool and the exosomes originated from antigen-presenting cells present in bronchoalveolar lavage fluid of asthmatic patients, respectively (103,104). Combinations of antibodies directed against different biomarkers have been also used (105) and the present effort is towards more systematic analyses, such as that allowed by using the multiplexed platform nPLEX (106). Although the survey was limited by the number and quality of available antibodies (by far lower than the device potential), it demonstrated that for each cell type and its corresponding exosomes there are combinations of right antibody/antigen pairs suitable for selective separation and diagnostics. Other EV stratification approaches based on multiple IA have been proposed (107) and they share the procedure depicted in Figure 2. Different EV classes were separated from the culture medium of endothelial and endothelial progenitor cells by using a pool of antibody-conjugated microbeads in combination with quantum-dots conjugated with a second set of specific antibodies (108). Quantum dot was also used to amplify the signal of another multi-antibody capture system which enabled the identification of as few as 100 exosomes/µL of tumor patient serum samples (109). In another experimental setting, gold-coated glass sensor chips were functionalized with antibody microarrays specific for the extracellular domains of exosome membrane proteins for monitoring the EV distribution by surface plasmon resonance imaging (110). IA of exosomes obtained by employing variable combinations of antibodies in a microfluidic system is reported also in He et al. (111) but the lack of detailed information relative to the used experimental methodology impairs to evaluate the quality of this contribution. Recently some new accessible membrane antigens have been proposed as biomarker candidates for prostate (gamma-glutamyltransferase 1, LAMTOR1, transmembrane protein 256) and pancreas (glypican-1) cancers (112-114). The task of successful exploitation remains not trivial since EV biomarkers can be slightly modified with respect to the structure of the homologue present in the original cell (115) or in related exosomes (116). Since this condition can impair the epitope recognition of conventional antibodies, new reagents should be probably developed.
In the perspective of obtaining the further stratification of the exosome fraction, the introduction of IA protocols that use new antigen/antibody pairs for the isolation of exclusive exosome groups is highly encouraging. A33 was initially identified as a biomarker suitable for targeting and immunocapture of exosomes released by human colon tumor cells (117). Successively, by applying sequential immunocapture with anti-A33 and anti-EpCAM to the same human colon carcinoma cell supernatant sample, it was possible to separate two exosome populations (putatively released from basolateral and apical cell surface, respectively) which shared only a minor fraction of their miRNA cargo between them and with the microvesicle fraction separated by differential centrifugation (49,118). The phosphorylation state of IRS-1 in neural derived exosomes was assessed after selective streptavidin-dependent immunoprecipitation of exosomes labelled with biotinylated antibodies against anti-neural cell adhesion molecule-1 (NCAM-1) and anti-neural cell adhesion molecule L1 (L1CAM/CD171) antibodies (119). L1CAM-dependent immunocapture was further successfully performed for recovering α-synuclein-containing exosomes released from central neuron cells (120). EVs from urine of kidney transplanted patients were sorted by using magnetic beads functionalized with anti-CD133 commercial antibodies and their presence allowed to monitor renal functionality (121). Similar magnetic beads functionalized with anti-CD34 antibodies enabled the recovery of specific AML blast-derived exosomes directly from patient plasma (122). Magnetic beads functionalized with monoclonal antibodies against complement receptor type 1 (CR1) were used to immunoprecipitate specifically podocyte derived-exosomes (123). Finally, IA performed using anti-CD45 has been exploited to remove hematopoietic exosome contaminants from virus particle samples (124). Other results cannot be easily assessed because of poor methodological description but indicate the growing interest in this research field (125,126).
Immunocapture is also the most used enrichment method for the recovery of small amounts of EVs for diagnostics goals in conventional set-ups (ELISA plates, magnetic beads and fluorescent beads for flow-cytometry), in microfluidic devices such as Exochip and ExoSearch (51,127), lateral flow immunoassay (LFIA) (128), and in monolithic silica micro-tips (129). These tools have been usually conceived to capture possibly any meaningful exosome present in a sample and therefore they exploit antibodies against antigens “universally” expressed in such vesicles, such as CD63Jeny (130-133), CD81 (128,134), CD9 (54,112,128,129), or against generic epithelial tumor markers such as EpCAM (133,135,136) and its combination with CD24 and CA-125 (127).
Comparative analyses
The different EV purification methods result in variable exosome yield and contamination levels. Recently it has also reported that they can induce the detachment of EV components form the vesicle surface (100). Several research groups evaluated the available protocols and commercial products to establish their reliability and to point out their critical aspects (50,54). When UC, OptiPrep™ DG separation, and IA capture using anti-EpCAM-coated magnetic beads (IA) were compared (44,137), it resulted that IA was the most efficient method to isolate exosome and exosome-associated proteins. It yielded at least twofold more material than the two other alternative methods as monitored by mass spectrometry assessing the enrichment of marker proteins associated with exosome biogenesis, function, sorting, intracellular trafficking, and internalization in a recipient cell. IA separation performed better than chemical-physical methods also in previous works (138). The drawback of the IA approach is its dependence on the availability of convenient antibodies. If this factor is limiting, the authors advise using DG centrifugation (137) or SEC (138). With respect to UC, DG yields are significantly lower in terms of total protein, but the exosome samples are not contaminated as those recovered after standard UC. OptiPrepTM DG centrifugation outperformed UC and both ExoQuickTM (EQ, System Biosciences, Palo Alto, CA, USA) (139) and Total Exosome IsolationTM (TEI, ThermoFisher, Bridgewater, NJ, USA) (140) precipitation methods also in another systematic assessment (52). Also in this case, the methods which provided higher yields in term of total protein (EQ and TEI) performed very badly in terms of exosome purity, as demonstrated by immune electron microscopy, western blot, RT-qPCR, and NTA. EQ and the peptide-based precipitation method exploiting the METM kit (141) as well as other commercial kits resulted producing the most contaminated material also in further comparative surveys which demonstrated the highest quality of the samples obtained by immunopurification (54,142). Apart from publications apparently directly sponsored by commercial companies, we found only one comparative survey in which precipitation kits were evaluated positively (143), but the authors used conventional UC as a reference instead of the more suitable protocols presently used by EV specialists (46,51).
Conclusions
The awareness about the necessity to have reproducible methods to purify exosome fractions not contaminated with other sample components as well as the interest to discriminate among distinct subpopulations to analyse separately their content supported innovative technical solutions for EV fractionation. Pre-clearing steps have been proposed to diminish sample complexity and disrupting undesired interactions with other biological components (144-146) and combined techniques, in which a final immunocapture enables high selectivity after cycles of (gradient) centrifugation, filtration and SEC, represent the state-of-the-art more than conventional multi-step centrifugation. In particular, since the medical and scientific interests moved to selectivity rather than larger yields, methods based on affinity and IA became more and more attractive and demonstrated to be sufficient for recovering highly purified exosomes even directly from complex biological media, without preliminary steps (97,120,123,129,147). In this perspective, the trend seems to be using combinations of biomarkers rather than single ones to identify more specifically distinct EV sub-populations (98,113) and separate them using different device configurations (106,128). The progressive identification of EV/exosome sub-group biomarkers could lead to the development of multi-parameter flow cytometry-based sorting of exosome/EV subpopulations (Figure 3) (148,149), in a way resembling the characterization and separation of hematopoietic cells at different degree of their differentiation. In this optic of multi-parametric labelling, non-exclusive, quantitative biomarkers could contribute to differentiate between exosome subgroups. The limiting factor could become the production of suitable antibodies able to recognize (enough) exosome-specific epitopes.

The conventional procedure would consider: (I) inferring potential exosome biomarkers by biology and proteomics results; (II) producing the corresponding recombinant proteins or their soluble domains; (III) evaluating their actual binding to EV epitopes. An alternative method is the blind direct antibody selection on exosomes to recover conformational-specific reagents. This approach is feasible in vitro panning pre-immune recombinant antibody libraries. We applied it successfully to recover nanobodies able to discriminate between very similar cell types (150) and obtained promising preliminary results panning directly on exosomes (Popovic et al., in preparation) (151). The resulting recombinant antibodies are inexpensive to produce and simple to engineer and their straightforward labelling (152) is compatible with multi-parameter flow cytometry.
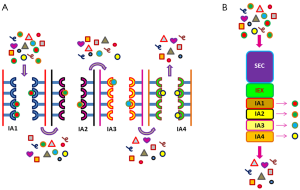
Another interesting development could consist in a combined chromatographic protocol in which SEC and IA are integrated with ion-exchange (Figure 4). In particular, monolith columns could be the material of choice because of their pore diameter can be selected according to the needs, the structure is preserved even at high pressure, and volumes are scalable. The reliability of this material for exosome purification was already demonstrated by the applications in which the capture antibodies were immobilized into monolithic tips (129) and EVs were recovered by mini-SEC (122). Columns formed by successive monolith discs with different chemical, physical, and affinity features (SEC + ion exchange + multiple IA chromatography) could be customized for the recovery of exosome subclasses.
Acknowledgments
Funding: This work was supported by the Slovenia-Serbia bilateral ARRS program (Project “Isolation of recombinant nanobodies specific for exosome subclasses”, BI-RS/16-17-014), by the Slovenia-Belgium ARRS-FWO program (Project “Identification of recombinant nanobodies for exosome immune-capture in breast cancer diagnosis”, ARRS/N4-0046-5100-1/2015-59) and by the Grant No. 172049 from Ministry of Education, Science and Technological Development of the Republic of Serbia.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.44). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet 2012;21:R125-34. [Crossref] [PubMed]
- EL Andaloussi S. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347-57. [Crossref] [PubMed]
- van der Pol E, Boing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 2012;64:676-705. [Crossref] [PubMed]
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 1983;33:967-78. [Crossref] [PubMed]
- Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur J Cell Biol 1984;35:256-63. [PubMed]
- Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 1996;183:1161-72. [Crossref] [PubMed]
- Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med 1998;4:594-600. [Crossref] [PubMed]
- Wahlgren J, Karlson TDL, Glader P, et al. Activated Human T Cells Secrete Exosomes That Participate in IL-2 Mediated Immune Response Signaling. PLoS One 2012;7:e49723 [Crossref] [PubMed]
- Fauré J, Lachenal G, Court M, et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci 2006;31:642-8. [Crossref] [PubMed]
- Guescini M, Genedani S, Stocchi V, et al. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna) 2010;117:1-4. [Crossref] [PubMed]
- Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006;20:847-56. [Crossref] [PubMed]
- Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med 2011;6:481-92. [Crossref] [PubMed]
- Mu J, Zhuang X, Wang Q, et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res 2014;58:1561-73. [Crossref] [PubMed]
- Nosanchuk JD, Nimrichter L, Casadevall A, et al. A role for vesicular transport of macromolecules across cell walls in fungal pathogenesis. Commun Integr Biol 2008;1:37-9. [Crossref] [PubMed]
- Oliveira DL, Nakayasu ES, Joffe LS, et al. Characterization of Yeast Extracellular Vesicles: Evidence for the Participation of Different Pathways of Cellular Traffic in Vesicle Biogenesis. PLoS One 2010;5:e11113 [Crossref] [PubMed]
- Chatterjee SN, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol 1967;49:1-11. [Crossref] [PubMed]
- Chaput N, Thery C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol 2011;33:419-40. [Crossref] [PubMed]
- Bobrie A, Colombo M, Raposo G, et al. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 2011;12:1659-68. [Crossref] [PubMed]
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009;9:581-93. [Crossref] [PubMed]
- Roberg-Larsen H, Lund K, Seterdal KE, et al. Mass spectrometric detection of 27-hydroxycholesterol in breast cancer exosomes. J Steroid Biochem Mol Biol 2017;169:22-8. [Crossref] [PubMed]
- Gatti S, Bruno S, Deregibus MC, et al. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrol Dial Transplant 2011;26:1474-83. [Crossref] [PubMed]
- Jaiswal R, Johnson MS, Pokharel D, et al. Microparticles shed from multidrug resistant breast cancer cells provide a parallel survival pathway through immune evasion. BMC Cancer 2017;17:104. [Crossref] [PubMed]
- Del Conde I, Shrimpton CN, Thiagarajan P, et al. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 2005;106:1604-11. [Crossref] [PubMed]
- Rak J, Guha A. Extracellular vesicles--vehicles that spread cancer genes. Bioessays 2012;34:489-97. [Crossref] [PubMed]
- Camussi G, Deregibus MC, Bruno S, et al. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res 2011;1:98-110. [PubMed]
- An T, Qin S, Xu Y, et al. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles 2015;4:27522. [Crossref] [PubMed]
- Mack M, Kleinschmidt A, Bruhl H, et al. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med 2000;6:769-75. [Crossref] [PubMed]
- Hosseini HM, Fooladi AA, Nourani MR, et al. The role of exosomes in infectious diseases. Inflamm Allergy Drug Targets 2013;12:29-37. [Crossref] [PubMed]
- Bellingham SA, Guo BB, Coleman BM, et al. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front Physiol 2012;3:124. [Crossref] [PubMed]
- Emmanouilidou E, Melachroinou K, Roumeliotis T, et al. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci 2010;30:6838-51. [Crossref] [PubMed]
- Vella LJ, Sharples RA, Lawson VA, et al. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol 2007;211:582-90. [Crossref] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Bruno S, Grange C, Collino F, et al. Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS One 2012;7:e33115 [Crossref] [PubMed]
- Herrera MB, Fonsato V, Gatti S, et al. Human liver stem cell-derived microvesicles accelerate hepatic regeneration in hepatectomized rats. J Cell Mol Med 2010;14:1605-18. [Crossref] [PubMed]
- Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother 2006;55:808-18. [Crossref] [PubMed]
- Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012;10:e1001450 [Crossref] [PubMed]
- Kim DK, Kang B, Kim OY, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles 2013;2. [PubMed]
- Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles 2012;1. [PubMed]
- Escrevente C, Keller S, Altevogt P, et al. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011;11:108. [Crossref] [PubMed]
- Gonzales PA, Pisitkun T, Hoffert JD, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 2009;20:363-79. [Crossref] [PubMed]
- Jeppesen DK, Nawrocki A, Jensen SG, et al. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics 2014;14:699-712. [Crossref] [PubMed]
- Østergaard O, Nielsen CT, Iversen LV, et al. Quantitative proteome profiling of normal human circulating microparticles. J Proteome Res 2012;11:2154-63. [Crossref] [PubMed]
- Revenfeld AL, Baek R, Nielsen MH, et al. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther 2014;36:830-46. [Crossref] [PubMed]
- Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012;56:293-304. [Crossref] [PubMed]
- Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3:26913. [Crossref] [PubMed]
- Van Deun J, Mestdagh P, Agostinis P, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods 2017;14:228-32. [Crossref] [PubMed]
- György B, Hung ME, Breakefield XO, et al. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol 2015;55:439-64. [Crossref] [PubMed]
- Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles 2013;2. [PubMed]
- Ji H, Chen M, Greening DW, et al. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS One 2014;9:e110314 [Crossref] [PubMed]
- Taylor DD, Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015;87:3-10. [Crossref] [PubMed]
- Xu R, Greening DW, Zhu HJ, et al. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest 2016;126:1152-62. [Crossref] [PubMed]
- Van Deun J, Mestdagh P, Sormunen R, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles 2014;3. [PubMed]
- Coumans FAW, Brisson AR, Buzas EI, et al. Methodological Guidelines to Study Extracellular Vesicles. Circ Res 2017;120:1632-48. [Crossref] [PubMed]
- Zarovni N, Corrado A, Guazzi P, et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015;87:46-58. [Crossref] [PubMed]
- Momen-Heravi F, Balaj L, Alian S, et al. Current methods for the isolation of extracellular vesicles. Biol Chem 2013;394:1253-62. [Crossref] [PubMed]
- György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011;68:2667-88. [Crossref] [PubMed]
- Cvjetkovic A, Lötvall J, Lässer C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles 2014;3. [PubMed]
- Momen-Heravi F, Balaj L, Alian S, et al. Impact of Biofluid Viscosity on Size and Sedimentation Efficiency of the Isolated Microvesicles. Front Physiol 2012;3:162. [Crossref] [PubMed]
- Tangney CC, Hafner JM, McQuiston BD, et al. Postprandial changes in plasma and serum viscosity and plasma lipids and lipoproteins after an acute test meal. Am J Clin Nutr 1997;65:36-40. [Crossref] [PubMed]
- Jeurissen S, Vergauwen G, Van Deun J, et al. The isolation of morphologically intact and biologically active extracellular vesicles from the secretome of cancer-associated adipose tissue. Cell Adh Migr 2017;11:196-204. [Crossref] [PubMed]
- Kalra H, Adda CG, Liem M, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013;13:3354-64. [Crossref] [PubMed]
- Cantin R, Diou J, Belanger D, et al. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J Immunol Methods 2008;338:21-30. [Crossref] [PubMed]
- Vergauwen G, Dhondt B, Van Deun J, et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci Rep 2017;7:2704. [Crossref] [PubMed]
- Li P, Kaslan M, Lee SH, et al. Progress in Exosome Isolation Techniques. Theranostics 2017;7:789-804. [Crossref] [PubMed]
- Palma J, Yaddanapudi SC, Pigati L, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res 2012;40:9125-38. [Crossref] [PubMed]
- Aalberts M, van Dissel-Emiliani FM, van Adrichem NP, et al. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol Reprod 2012;86:82. [Crossref] [PubMed]
- Yuana Y, Levels J, Grootemaat A, et al. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles 2014;3. [PubMed]
- Sódar BW, Kittel Á, Pálóczi K, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep 2016;6:24316. [Crossref] [PubMed]
- Sodi R, Eastwood J, Caslake M, et al. Relationship between circulating microRNA-30c with total- and LDL-cholesterol, their circulatory transportation and effect of statins. Clin Chim Acta 2017;466:13-9. [Crossref] [PubMed]
- Quintana JF, Makepeace BL, Babayan SA, et al. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit Vectors 2015;8:58. [Crossref] [PubMed]
- Zeringer E, Barta T, Li M, et al. Strategies for isolation of exosomes. Cold Spring Harb Protoc 2015;2015:319-23. [PubMed]
- Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release 2015;219:396-405. [Crossref] [PubMed]
- Cheruvanky A, Zhou H, Pisitkun T, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol 2007;292:F1657-61. [Crossref] [PubMed]
- Escudier B, Dorval T, Chaput N, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med 2005;3:10. [Crossref] [PubMed]
- Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med 2005;3:9. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer 2005;92:305-11. [Crossref] [PubMed]
- Deregibus MC, Figliolini F, D'antico S, et al. Charge-based precipitation of extracellular vesicles. Int J Mol Med 2016;38:1359-66. [Crossref] [PubMed]
- Kim DK, Nishida H, An SY, et al. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc Natl Acad Sci U S A 2016;113:170-5. [Crossref] [PubMed]
- Buzzi D, Corrado A, Jančar J, et al. Development of a separation method for exosomes by means of large pores monolithic supports. 2014. Available online: http://www.biaseparations.com/support/posters/product/775-development-of-a-separation-method-for-exosomes-by-means-of-large-pore-monolithic-columns
- Steppert P, Burgstaller D, Klausberger M, et al. Purification of HIV-1 gag virus-like particles and separation of other extracellular particles. J Chromatogr A 2016;1455:93-101. [Crossref] [PubMed]
- Oh S, Kang DJ, Ahn SM, et al. Miniaturized asymmetrical flow field-flow fractionation: Application to biological vesicles. J Sep Sci 2007;30:1082-7. [Crossref] [PubMed]
- Kang D, Oh S, Ahn SM, et al. Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography-tandem mass spectrometry. J Proteome Res 2008;7:3475-80. [Crossref] [PubMed]
- Ashby J, Flack K, Jimenez LA, et al. Distribution Profiling of Circulating MicroRNAs in Serum. Anal Chem 2014;86:9343-9. [Crossref] [PubMed]
- Petersen KE, Manangon E, Hood JL, et al. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal Bioanal Chem 2014;406:7855-66. [Crossref] [PubMed]
- Sitar S, Kejzar A, Pahovnik D, et al. Size Characterization and Quantification of Exosomes by Asymmetrical-Flow Field-Flow Fractionation. Anal Chem 2015;87:9225-33. [Crossref] [PubMed]
- Agarwal K, Saji M, Lazaroff SM, et al. Analysis of Exosome Release as a Cellular Response to MAPK Pathway Inhibition. Langmuir 2015;31:5440-8. [Crossref] [PubMed]
- Yang JS, Lee JC, Byeon SK, et al. Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal Chem 2017;89:2488-96. [Crossref] [PubMed]
- Inc SB. ExoQuick overview. Available online: https://www.systembio.com/microrna-research/exoquick-exosomes/overview
- Alvarez ML, Khosroheidari M, Kanchi Ravi R, et al. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int 2012;82:1024-32. [Crossref] [PubMed]
- Lau C, Kim Y, Chia D, et al. Role of Pancreatic Cancer-derived Exosomes in Salivary Biomarker Development. J Biol Chem 2013;288:26888-97. [Crossref] [PubMed]
- Echevarria J, Royo F, Pazos R, et al. Microarray-based identification of lectins for the purification of human urinary extracellular vesicles directly from urine samples. Chembiochem 2014;15:1621-6. [Crossref] [PubMed]
- Balaj L, Atai NA, Chen W, et al. Heparin affinity purification of extracellular vesicles. Sci Rep 2015;5:10266. [Crossref] [PubMed]
- Yáñez-Mó M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:27066. [Crossref] [PubMed]
- Purushothaman A, Bandari SK, Liu J, et al. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J Biol Chem 2016;291:1652-63. [Crossref] [PubMed]
- Nakai W, Yoshida T, Diez D, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep 2016;6:33935. [Crossref] [PubMed]
- Lai RC, Tan SS, Yeo RW, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J Extracell Vesicles 2016;5:29828. [Crossref] [PubMed]
- Reiner AT, Tan S, Agreiter C, et al. EV-Associated MMP9 in High-Grade Serous Ovarian Cancer Is Preferentially Localized to Annexin V-Binding EVs. Dis Markers 2017;2017:9653194
- Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods 2001;247:163-74. [Crossref] [PubMed]
- Koga K, Matsumoto K, Akiyoshi T, et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res 2005;25:3703-7. [PubMed]
- McBride JD, Menocal-Rodriguez L, Guzman W, et al. Bone marrow mesenchymal stem cell-derived CD63+ exosomes transport Wnt3a exteriorly and enhance dermal fibroblast proliferation, migration and angiogenesis in vitro. Stem Cells Dev 2017;26:1384-98. [Crossref] [PubMed]
- Caby MP, Lankar D, Vincendeau-Scherrer C, et al. Exosomal-like vesicles are present in human blood plasma. Int Immunol 2005;17:879-87. [Crossref] [PubMed]
- Bukong TN, Momen-Heravi F, Kodys K, et al. Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90. PLoS Pathog 2014;10:e1004424 [Crossref] [PubMed]
- Wubbolts R, Leckie RS, Veenhuizen PTM, et al. Proteomic and biochemical analyses of human B cell-derived exosomes - Potential implications for their function and multivesicular body formation. J Biol Chem 2003;278:10963-72. [Crossref] [PubMed]
- Levänen B, Bhakta NR, Torregrosa Paredes P, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol 2013;131:894-903. [Crossref] [PubMed]
- Kadiu I, Narayanasamy P, Dash PK, et al. Biochemical and Biologic Characterization of Exosomes and Microvesicles as Facilitators of HIV-1 Infection in Macrophages. J Immunol 2012;189:744-54. [Crossref] [PubMed]
- Im H, Shao HL, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32:490-5. [Crossref] [PubMed]
- Hildonen S, Skarpen E, Halvorsen TG, et al. Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci Rep 2016;6:36331. [Crossref] [PubMed]
- Wang J, Guo R, Yang Y, et al. The Novel Methods for Analysis of Exosomes Released from Endothelial Cells and Endothelial Progenitor Cells. Stem Cells Int 2016;2016:2639728
- Boriachek K, Islam MN, Gopalan V, et al. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst 2017;142:2211-9. [Crossref] [PubMed]
- Zhu L, Wang K, Cui J, et al. Label-Free Quantitative Detection of Tumor-Derived Exosomes through Surface Plasmon Resonance Imaging. Anal Chem 2014;86:8857-64. [Crossref] [PubMed]
- He M, Crow J, Roth M, et al. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014;14:3773-80. [Crossref] [PubMed]
- Kawakami K, Fujita Y, Matsuda Y, et al. Gamma-glutamyltransferase activity in exosomes as a potential marker for prostate cancer. BMC Cancer 2017;17:316. [Crossref] [PubMed]
- Øverbye A, Skotland T, Koehler CJ, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015;6:30357-76. [Crossref] [PubMed]
- Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82. [Crossref] [PubMed]
- Taylor DD, Homesley HD, Doellgast GJ. Binding of Specific Peroxidase-Labeled Antibody to Placental-Type Phosphatase on Tumor-Derived Membrane-Fragments. Cancer Res 1980;40:4064-9. [PubMed]
- Shao H, Chung J, Lee K, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun 2015;6:6999. [Crossref] [PubMed]
- Mathivanan S, Lim JW, Tauro BJ, et al. Proteomics Analysis of A33 Immunoaffinity-purified Exosomes Released from the Human Colon Tumor Cell Line LIM1215 Reveals a Tissue-specific Protein Signature. Mol Cell Proteomics 2010;9:197-208. [Crossref] [PubMed]
- Tauro BJ, Greening DW, Mathias RA, et al. Two Distinct Populations of Exosomes Are Released from LIM1863 Colon Carcinoma Cell-derived Organoids. Mol Cell Proteomics 2013;12:587-98. [Crossref] [PubMed]
- Kapogiannis D, Boxer A, Schwartz JB, et al. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer's disease. FASEB J 2015;29:589-96. [Crossref] [PubMed]
- Shi M, Liu CQ, Cook TJ, et al. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson's disease. Acta Neuropathologica 2014;128:639-50. [Crossref] [PubMed]
- Dimuccio V, Ranghino A, Praticò Barbato L, et al. Urinary CD133+ extracellular vesicles are decreased in kidney transplanted patients with slow graft function and vascular damage. PLoS One 2014;9:e104490 [Crossref] [PubMed]
- Hong CS, Muller L, Boyiadzis M, et al. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One 2014;9:e103310 [Crossref] [PubMed]
- Prunotto M, Farina A, Lane L, et al. Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J Proteomics 2013;82:193-229. [Crossref] [PubMed]
- Coren LV, Shatzer T, Ott DE. CD45 immunoaffinity depletion of vesicles from Jurkat T cells demonstrates that exosomes contain CD45: no evidence for a distinct exosome/HIV-1 budding pathway. Retrovirology 2008;5:64. [Crossref] [PubMed]
- Rana S, Yue SJ, Stadel D, et al. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol 2012;44:1574-84. [Crossref] [PubMed]
- Redzic JS, Kendrick AA, Bahmed K, et al. Extracellular vesicles secreted from cancer cell lines stimulate secretion of MMP-9, IL-6, TGF-beta1 and EMMPRIN. PLoS One 2013;8:e71225 [Crossref] [PubMed]
- Zhao Z, Yang Y, Zeng Y, et al. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016;16:489-96. [Crossref] [PubMed]
- Oliveira-Rodríguez M, López-Cobo S, Reyburn HT, et al. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J Extracell Vesicles 2016;5:31803. [Crossref] [PubMed]
- Ueda K, Ishikawa N, Tatsuguchi A, et al. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci Rep 2014;4:6232. [Crossref] [PubMed]
- Wei F, Yang J, Wong DT. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens Bioelectron 2013;44:115-21. [Crossref] [PubMed]
- Yang J, Wei F, Schafer C, et al. Detection of Tumor Cell-Specific mRNA and Protein in Exosome-Like Microvesicles from Blood and Saliva. PLoS One 2014;9:e110641 [Crossref] [PubMed]
- Kanwar SS, Dunlay CJ, Simeone DM, et al. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014;14:1891-900. [Crossref] [PubMed]
- Fang S, Tian H, Li X, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One 2017;12:e0175050 [Crossref] [PubMed]
- Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip 2016;16:3033-42. [Crossref] [PubMed]
- Kim G, Yoo CE, Kim M, et al. Noble Polymeric Surface Conjugated with Zwitterionic Moieties and Antibodies for the Isolation of Exosomes from Human Serum. Bioconjug Chem 2012;23:2114-20. [Crossref] [PubMed]
- Yoo CE, Kim G, Kim M, et al. A direct extraction method for microRNAs from exosomes captured by immunoaffinity beads. Anal Biochem 2012;431:96-8. [Crossref] [PubMed]
- Greening DW, Xu R, Ji H, et al. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol 2015;1295:179-209. [Crossref] [PubMed]
- Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol 2011;728:235-46. [Crossref] [PubMed]
- Peterson MF, Otoc N, Sethi JK, et al. Integrated systems for exosome investigation. Methods 2015;87:31-45. [Crossref] [PubMed]
- Li M, Rai AJ, DeCastro GJ, et al. An optimized procedure for exosome isolation and analysis using serum samples: Application to cancer biomarker discovery. Methods 2015;87:26-30. [Crossref] [PubMed]
- Ghosh A, Davey M, Chute IC, et al. Rapid Isolation of Extracellular Vesicles from Cell Culture and Biological Fluids Using a Synthetic Peptide with Specific Affinity for Heat Shock Proteins. PLoS One 2014;9:e110443 [Crossref] [PubMed]
- Brett SI, Lucien F, Guo C, et al. Immunoaffinity based methods are superior to kits for purification of prostate derived extracellular vesicles from plasma samples. Prostate 2017;77:1335-43. [Crossref] [PubMed]
- Helwa I, Cai J, Drewry MD, et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS One 2017;12:e0170628 [Crossref] [PubMed]
- Gallart-Palau X, Serra A, Wong AS, et al. Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR). Sci Rep 2015;5:14664. [Crossref] [PubMed]
- Mørk M, Handberg A, Pedersen S, et al. Prospects and limitations of antibody-mediated clearing of lipoproteins from blood plasma prior to nanoparticle tracking analysis of extracellular vesicles. J Extracell Vesicles 2017;6:1308779 [Crossref] [PubMed]
- Puhka M, Nordberg ME, Valkonen S, et al. KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. Eur J Pharm Sci 2017;98:30-9. [Crossref] [PubMed]
- Hong CS, Funk S, Muller L, et al. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles 2016;5:29289. [Crossref] [PubMed]
- Volgers C, Benedikter BJ, Grauls GE, et al. Bead-based flow-cytometry for semi-quantitative analysis of complex membrane vesicle populations released by bacteria and host cells. Microbiol Res 2017;200:25-32. [Crossref] [PubMed]
- Hauser P, Wang S, Didenko VV. Apoptotic Bodies: Selective Detection in Extracellular Vesicles. Methods Mol Biol 2017;1554:193-200. [Crossref] [PubMed]
- Crépin R, Gentien D, Duché A, et al. Nanobodies against surface biomarkers enable the analysis of tumor genetic heterogeneity in uveal melanoma patient-derived xenografts. Pigment Cell Melanoma Res 2017;30:317-27. [Crossref] [PubMed]
- Popovic M, Toffoleto B, Casseli D, et al. Successful panning of pre-immune VHH phage library directly on exosomes – FEBS3+ Meeting Molecules of Life, Portoroz, Slovenija, 2015.
- Djender S, Schneider A, Beugnet A, et al. Bacterial cytoplasm as an effective cell compartment for producing functional VHH-based affinity reagents and Camelidae IgG-like recombinant antibodies. Microb Cell Fact 2014;13:140. [Crossref] [PubMed]


