
Research progress on DNA methylation in hepatocellular carcinoma
Liver cancer (also known as hepatic cancer or primary hepatic cancer) is a malignant cancer originating in the liver which severely affects public health and quality of life. Globally, liver cancer is the 5th most common cancer in male patients and the seventh most common in female patients. The majority of liver cancer cases occur in the developing countries, especially in Southeast Asia and sub-Saharan Africa where there is a high prevalence of hepatitis B virus (HBV) infection (1).
Hepatocellular carcinoma (HCC) and epigenetics
HCC is the most common histological subtype of liver cancer, accounting for 70–85% of total liver cancer cases (2,3). In 2008, around 700,000 HCC cases were diagnosed worldwide (4). In the past 20 years, the HCC incidence in the U.S. increased by three times with a 5-year survival rate lower than 12%, and HCC related to persistent hepatitis C virus (HCV) infection has become fastest-growing cause of cancer-related death (1).
In China, HCC is the third most common cause of cancer-related death in both male and female patients (5). According to previous studies, the inhibited growth rate of HCC in China is attributable to several different advances in public health, including effective control of contagious HBV and HCV infection, reduction of food contamination by aflatoxin, improved quality of drinking water, and medical injection procedures which have been more standardized and safer. After the Chinese government initiated HBV vaccination to infants and children in areas with high risk of HCC in 1986, fatality due to HCC decreased by 95% over the following 15 years among those aged 0–19 (6-10). However, the overall incidence of HCC is still rising, due to various factors including unhealthy lifestyle (smoking, heavy drinking, input of high-energy food), and non-alcoholic fatty liver disease.
The pathogenesis of HCC involves interaction between environmental and genetic factors (1,11,12). The major factors in HCC development include HBV infection, HCV infection, alcohol abuse, non-alcoholic fatty liver disease, and exposure to environmental carcinogens (e.g., aflatoxin B1). Among the less common factors are hereditary hemochromatosis, α1-antitrypsin deficiency, autoimmune hepatitis, hematoporphyrinism, and Wilson’s disease (1,13). The precise molecular mechanisms of HCC are very complicated and heterogeneous, impeding clinical diagnosis and treatment (3).
To date, the main issues with HCC treatment are insufficient early detection and rapid tumor development. Despite the fact that clinical and imaging criteria for early HCC diagnosis have been available, and that surgical dissection and liver transplant have been commonly performed as mature therapeutic approaches, the sensitivity and specificity of existing tumor biomarker tests (e.g., Alpha-fetoprotein) are still relatively low when screening for HCC. Most patients are diagnosed and treated at later stages of HCC, which greatly limits the selection of available treatments and leads to poor prognoses (3,14-16). In addition, due to patient variability in tumor development rates, HCC patients receiving the same treatment may exhibit different prognoses and survival rates. Reliable prognosis prediction and classification/categorization cannot be obtained based on current diagnostic and evaluating methods (17). Therefore, improving early detection can facilitate the selection of treatment options, which may help to achieve better clinical outcomes for HCC patients.
In 1942, Waddington discovered the phenomenon of inconsistency between genotype and phenotype, giving rise to the discipline of epigenetics which plays a critical role in the regulation of gene expression (18). The key aspects of epigenetics include DNA methylation, genomic imprinting, maternal effects, gene silencing, nucleolar dominance, activation of dormant transposons, and RNA editing. Epigenetics studies address how environmental factors interact with the genome and generate heritable modification of gene expression and cellular phenotype without altering DNA sequence. Epigenetic modification can be affected by cellular metabolism and extracellular environments, such as pathogenic factors. Therefore, studying epigenetics is critical to understanding the mechanisms underlying the pathogenesis and progression of human diseases (17-19).
DNA methylation abnormalities, which play an important role in the onset of tumor development (19-28), have been observed in multiple cancers including HCC. According to several studies, abnormal DNA methylation can be used as a molecular biomarker for tumor screening and prognosis prediction (29-32). Screening for DNA methylation abnormalities, in combination with integrated analysis of gene expression, can be used to identify the epigenetic events driving tumorigenesis (33,34). Several groups have reported that the of the unique and specific arrangement of DNA methylation abnormalities is correlated to HCC diagnosis and prognosis (13,28,35-38). Regional DNA hypermethylation is also known to occur during the early stages of HCC; thus, studying the association between DNA methylation levels and HCC staging may be useful for clinical diagnosis and prediction (39).
Emerging techniques for examination of targeted genome regions/loci
Three known kinds of DNA methylation modification are 5-methylcytosine (5mC), present in eukaryotes; N6-methyladenosine, mainly seen in bacteria and pathogenic organisms; and 7-methylguanine, which rarely occurs. In vivo formation of 5mC is catalyzed by DNA methyltransferases (DNMTs) using S-adenosyl methionine as a methyl provider, and selectively attaching a methyl group to the 5th carbon atom of the pyrimidine ring of cytosine (40). In eukaryotes the majority of 5mC formation occurs in the genomic regions that are abundant with cytosine-phosphate-guanine dinucleotides, also known as CpG islands. CpG islands are characterized by high CG percentages (usually >50%) and 200–500 base pairs.
Examination of DNA methylation can be divided into several categories: global DNA methylation analysis targeting the whole genome, gene-specific or locus-specific DNA methylation analysis, and methylation pattern/profiling analysis. Different approaches and techniques provide DNA methylation information on different levels, depending upon the goals of examination (41).
Techniques for global DNA methylation analysis include high-performance liquid chromatography (HPLC) (42), high-performance capillary electrophoresis (HPCE) (43), thin-layer chromatography (TLC) (44), methyl group acceptance assay (45), chloroacetaldehyde assay (46), and immunochemical analysis (47).
Techniques for gene-specific DNA methylation analysis include methylation-sensitive restriction enzyme-Southern (MSRE-Southern) (48), methylation-sensitive restriction endonuclease-PCR (49,50), methylation-specific PCR (51), methylation-sensitive single-nucleotide primer extension (52), direct bisulfite genomic sequencing (49), methylight PCR (53,54), methylation-specific single-strand conformation analysis (55-57), methylation-specific denaturing gradient gel electrophoresis (58), methylation-specific melting curve analysis (59), methylation-specific denaturing high-performance liquid chromatography (60), and methylation-specific oligonucleotide microarray (61).
Methylation pattern/profiling analysis includes restriction landmark genomic scanning (62), methylated CpG island amplification (63)-representational difference analysis (64) methylation-sensitive arbitrarily primed PCR (65), and differential methylation hybridization (66,67).
Traditional methods of HCC methylation analysis target specific genes and loci (e.g., oncogenes and tumor suppressor genes) and quantitatively measure methylation levels (68-70). For example, in combined bisulfite restriction analysis (COBRA), researchers process DNA samples with sodium bisulfite, amplify target sequences with PCR, and digest PCR products with specific restriction enzymes. Electrophoresis, probe hybridization, and quantitative scanning are then used to obtain the methylation ratio in the tested samples. Gene methylation levels can be determined from analysis of PCR bands, as methylated bands cannot be digested by restriction enzymes (71). The COBRA technique has been widely used for DNA methylation screening due to its many strengths—simplicity, efficiency, high compatibility, and low requirements for sample sequences availability. However, the constraints imposed by specific restriction enzyme sites do not allow for determination of complete genomic DNA methylation signature. Gene-specific or locus-specific DNA methylation analysis can provide methylation information about specific sites known to be associated with HCC. Methylation analysis, in combination with integrated analysis of gene expression, may also facilitate exploration of potential gene functions and possible epigenetic cancer drivers.
According to a study using a bisulfite sequencing technique to measure methylation levels in HCV-infected liver tissues, the promoter regions of two Wnt inhibitor genes—SFRP2 and DKK1—were hypermethylated in precancerous stages of chronic HCV infection and cirrhosis, which led to the silencing of Wnt and eventually the induction of HCC. The methylation level of the SFRP2 promoter region was highest in HCC, followed by cirrhosis and chronic hepatitis, and lowest in normal liver (72).
Nishida et al. performed methylation-specific PCR and observed the epigenetic inactivation of some tumor suppressor genes (HIC1, GSTP1, SOCS1, RASSF1, CDKN2A, APC, RUNX3, PRDM2) in the early stages of HCC. This specific subset of tumor suppressor genes was significantly hypermethylated, and their methylation frequencies were associated with time-to-HCC occurrence in chronic hepatitis C patients (CHC). Thus, the altered methylation of these genes may play a critical role in the early steps of carcinogenesis, and can provide potential prognostic value for determining the risk of CHC developing into HCC (68). Some tumor suppressor genes (CASP8, MINT31, PTGS2, CACNA1G) were found to present unaltered methylation levels in the early steps of HCC, but continuously increased methylation along with the progression of HCC. In contrast, another seven tumor suppressor genes (RASSF2, MINT1, MINT2, RPRM, SFRP2, CDH1, DCC) were merely hypermethylated in the highly aggressive (high grade) stages of HCC, suggesting that different tumor suppressor genes function at different stages or times of HCC development (68). Although there have been many attempts to study the abnormal DNA methylation on different gene sites, the biological effects of methylation in HCC development remain to be stringently evaluated. Furthermore, some abnormal methylations may be generated in response to unrelated events, rather than being actual tumor drivers (73). In the study by Nishida et al., the methylation levels of tumor suppressor genes paralleled expression levels, indicating the functional significance of methylation in tumor suppressor genes (68).
Several previous studies have reported that the expression levels of oncogenes (c-Myc, cyclinD1, β-catenin) and tumor suppressor genes (P16INK4A, P53, E-cadherin, DLC-1, pRb) are decreased to different extents in HCC (74). In human HCC the ubiquitous activation of Ras and JAK/STAT signaling pathways may be caused by the coincident abnormal methylation of inhibitors of Ras and JAK/STAT (75). Using methylation-specific PCR to analyze the promoter regions of 105 genes, Calvisi et al. observed the epigenetic silencing of Ras pathway inhibitors (SOCS-4,5, SPRY1, SPRY2, DAB2) and activation of Ras gene and its downstream genes (ERK, AKT, RAL). The down-regulation of the Ras inhibitors SPRY1, SPRY2, DAB2, SOCS4, SOCS5 and angiogenesis inhibitors BNIP3, BNIP3L, IGFBP3, EGLN2 were shown to be correlated with poor prognoses in HCC patients, indicating a relationship between site-specific DNA hypomethylation and tumor development (76). At the same time, the Ras signaling pathway promotes tumor angiogenesis through activating transcription of target genes—VEGF, HIF-1α, IL-8 (77,78). The expression levels of Ras and its downstream effectors are highest in HCC cells, followed by para-tumor tissues, and lowest in normal liver, suggesting important roles in the induction of angiogenesis in human HCC. Meanwhile, the DNA methylation index in HCC patients is inversely correlated with survival period, linking the consistently increased methylation of promoter regions in tumor suppressor genes with the progression of HCC (see Table 1).

Full table
Another important effector of HCC development and progression is tissue inhibitor of metalloproteinase-3 (TIMP-3), which is a matrix metalloproteinase (MMP) that can inhibit cellular growth and migration and prevent the formation of vasculature. TIMP-3 plays important roles in multiple cellular physiological processes including cell proliferation, migration, differentiation, angiogenesis, apoptosis, and so forth. Mannello et al. found that the hypermethylation of CpG islands of the TIMP-3 sequence in HCC patients caused its downregulation through epigenetic inactivation, and facilitated the metastasis of cancer cells (109).
The LINE-1 retrotransposon composes approximately 17% of the human genome, and its methylation level imposes great influence on global genomic methylation (110,111). Previous literature has suggested that hypomethylation of LINE-1 is closely related to poor prognoses in different cancers, including cancers of the colon, esophagus, stomach, and ovary (112-115). Several recent studies have discovered the significant hypomethylation of promoter regions of LINE-1 in the early stages of HCC (111,112,116-118). The activation of LINE-1 may be mediated by c-Met activation. Activated LINE-1 not only induces HCC, but is also significantly associated with poor prognosis, and may therefore be used as a potential biomarker to evaluate the clinical progress of HCC patients (112,116-118). Similarly, hypomethylation has been found in imprint gene regions, Alu sequences also exhibit active DNA demethylation in early steps of HCC (111). In HCC patients the hypermethylated H-cadherin gene becomes inactivated, and the hypomethylated vimentin gene becomes upregulated, leading to the assumption that the interaction of H-cadherin and vimentin prompts the onset and development of HCC (113).
At present there are several reports in the literature of atypical methylation patterns, global hypomethylation, or hypermethylation on specific gene promoter regions in HCC tumor cells. The hypermethylation of CpG islands in the promoter regions of tumor suppressor genes leads to transcription silencing and genomic instability (119-126). Calvisi et al. evaluated the methylation levels of HCC samples by assessing the incorporation of [3H]dCTP into DNA, and found that the extent of genome-wide hypomethylation and CpG hypermethylation correlates with the biological features and clinical outcomes of HCC patients (76).
High-throughput sequencing techniques
Advances in chip-based examination techniques
Along with the rapid development of high-throughput biology techniques, high-density DNA methylation array has become more and more widely used in HCC studies. Most previous studies have focused on only one or a few genes, the most comprehensive of which tested as many as 105 genes (69,76,104,127-132). Aided by the new generation sequencing techniques, however, HCC methylation studies transitioned from single-gene/site methylation analysis to global/genomic methylation analysis. Moreover, associated statistical analysis methods—hierarchical cluster analysis, principal components analysis, etc.—are capable of deriving further useful information.
Recently, Illumina chip have been used to study the DNA methylation patterns in various HCC models using diverse sample sizes, examination techniques, and etiological factors. A study using Illumina 1500 Golden Gate arrays and methylation-specific PCR found abnormal methylation on CpG promoter regions of 24 new genes as potential methylation markers for HCC; 4 of these genes (TNFRSF10C, HOXA9, NPY, and IRF5) were hypermethylated and subsequently down-regulated in the tumor tissues, and thus can be used as HCC biomarkers (90).
Another study using similar techniques discovered DNA methylation signatures associated with pathogenic risk factors, tumor progression, diagnosis, and prognosis of HCC. Abnormal modes of methylation were defined as hypermethylation of promoter regions and hypomethylation of non-promoter regions. DNA methylation of 124 CpG sites from 94 genes showed significant differences between tumor tissues and para-tumor tissues, with 34 genes being hypermethylated (including RASSF1, APC, and CDKN2A) and 90 being hypomethylated (including GABRA5, NOTCH4, and PGR). These abnormally methylated genes are involved in multiple critical cellular activities: apoptosis (IRAK3), immunoresponse (GSTM2), cell cycle regulation (CCND2), etc. Hypomethylation of imprint genes (GABRA5, GABRG3, ZIM3) was also reported in this study. Some of the CpG sites found were associated with pathogenic risk factors of HCC, as specifically indicated in this study: HBV infection (NAT2, VSPG2, DCC, NTKR3, etc.), HCV infection (RIK, CHGA), and alcohol consumption (DIO3, STAT5A) (28).
Hernandez-Vargas et al. used Illumina Golden Gate BeadArray technology to examine 30 HCC samples with HBV or HCV infection, and found that HCC tumor cells exhibited special DNA methylation patterns. Meanwhile, DNA methylation states were closely associated with major infection factors and tumor progression. Of all 1,505 CpG islands that were examined, those in the promoter regions of the APC, RASSF1A, CDHN2A and FZD7R3 genes showed significantly different methylation states between tumor tissue and para-tumor (normal) tissue. In addition, methylation of the DNMT1 promoter was linked to the low differentiation level of tumor (28).
One of the representative methylation arrays, the Infinium Human Methylation 27K beadchip, is widely used to quantitatively measure the methylation levels at different sites in the genome, and can cover 27,578 CpG sites from 14,495 genes. These genes include RefSeq genes from the NCBI CCDS database, oncogenes with high methylation frequency, tumor-associated target genes, and promoters of miRNAs (133). During Infinium Human Methylation 27K beadchip analysis, DNA samples undergo bisulfite salt treatment, whole genome amplification, hybridization and single-base amplification, fluorescent staining, and chip scanning before acquisition of DNA methylation data. Several studies have affirmed the reliability and comparability of chip-based methylation analysis and traditional pyrosequencing-based methylation analysis (36,134-136). Compared to classical single-gene/site methylation analysis, chip-based methylation analysis exhibits great advantages due to its high methylation detection capacity, absence of PCR steps, and its integration of both gene expression and microRNA profiling. As such, examining methylation status in HCC is not limited to researching methylation abnormality on specific well-known sites, but extends to massive exploration of the methylation distribution of CpG sites. The chip-based methylation analysis is therefore of great importance to our understanding of the whole picture of epigenetics in HCC and the disclosure of additional critical methylation sites.
Ammerpohl et al. used Illumina Human Methylation 27K array to analyze 27,578 CpG sites and found unique methylation patterns in promoter regions, CpG islands, and PRC2-targets in cases of cirrhosis and HCC. Compared to normal liver tissues, HCC cells had 998 CpG sites with hypomethylation and 278 CpG sites with hypermethylation. The genes corresponding with abnormal methylation were involved in cellular immunity, communication, signaling transduction, and cell movements. Compared to cirrhotic tissues, HCC had 1,050 CpG sites with hypomethylation and 378 CpG sites with hypermethylation, and corresponding genes were involved in maintenance of cellular structure, signaling pathways, cell movements, and immunity. The hypermethylated signaling pathway components in the study included heterogeneous trimeric G protein pathways (Giα and Gsα regulatory pathways), PI3K pathway, TNF-β pathway, etc.; the hypomethylated pathways included angiogenesis signaling pathway, acetylcholinergic M1&3 pathway, TNF-β and EGF receptor pathway. Specifically, eight sites located within eight genes (SPRR3, TNFSF15, ALOX12, ANGPTL7, CELSR1, CRMP1, GNRH2 and LOC55908) showed significantly different methylation levels between HCC, cirrhosis, and normal liver tissues (79).
One large-scale research project using the Illumina Human Methylation 27K array discovered that a total of 2,324 CpG sites significantly differed in methylation level between HCC tumor and non-tumor tissues, with 684 CpG sites (covering 548 genes) significantly hypermethylated and 1,640 (covering 1,290 genes) hypomethylated in tumor tissue. DAB2IP, BMP4, ZFP41. SPDY1, and CDKN2A were the most hypermethylated genes, while CCL20, AKT3, SCGB1D1, WFDC6, and PAX4 were the most hypomethylated genes. Within the tumor tissues, the majority of CpG islands showed significant hypermethylation, and most non-CpG sites were significantly hypomethylated, regardless of their proximity to promoter regions. Importantly, this study identified abnormal methylation in two novel pathways (Wnt and 5-HT4-type receptor-mediated signaling), as well as finding ten cellular pathways that overlapped with those from a previous study by Ammerpohl et al. (79).
Designed as an improved chip-based methylation sequencing technique, the Infinium Human Methylation 450K Beadchip (abbreviated as 450K beadchip) can cover more than 480,000 CpG sites, including 99% of RefSeq genes, 96% of CpG islands in the UCSC database, 92% of CpG shores (2 kb areas around CpG islands) and 86% of CpG shelves (2–4 kb away from CpG islands), HMM islands, FANTOM 4 activators, differentially methylated regions (DMRs), informatically predicted enhancers, DNase-sensitive sites, Ensemble regulatory features, loci in MHC regions, non-CpG methylation sites in human stem cells, as well as 90% sites from 27K beadchips (137). This 450K chip can be used to perform quantitative methylation analysis for single CpG sites, and also to provide a suitable platform for genome-wide association study (GWAS). The genomic regions with low density of CpG islands may be easily missed by traditional methylation sequencing, but can be detected with the 450K chip.
Compared with the 27K beadchip, using the 450K chip to examine global DNA methylation can help to uncover more sites with abnormal methylation, and simultaneously oversee the methylation distribution of whole genome.
DNA methylation changes in HCC have been confirmed by a large number of researchers, especially on the level of the whole genome (20,36,68,97,105). One large-scale study analyzed the global DNA methylation data from 800 HCC patients using the 450K beadchip and verified the presence of global DNA hypomethylation (GDH) or CpG island methylator phenotypes (CIMP) in around 90% of HCC samples (35). Another study reported the finding of 28,017 CpG sites with hypermethylation and 102,495 CpG sites with hypomethylation by comparing the DNA methylation pattern between HCC tissue and para-tumor tissues from 66 patients. None of the genes exhibited simultaneous abnormal hypermethylation and hypomethylation, implying that these two epigenetic events are independent biological mechanisms (13). Due to the extensive genome coverage available with the 450K beadchip, this study also explored the lesser-known genomic regions referred to as CpG shores and shelves. While most of the significantly hypermethylated CpG sites were located in CpG islands, the majority of the significantly hypomethylated CpG sites sit in open sea regions, shores (2 kb area around CpG island), and shelves (2–4 kb away from CpG island) (13). Similar observations have been made for other tumors (e.g., colorectal cancer), indicating that the location of abnormal methylation in CpG islands, shores, and shelves may be relevant to the onset of tumorigenesis (138-140).
Methylation beadchip analysis can also reveals pecificin formation about etiological or risk factors in HCC patients. According to a study by Shen et al. that tested liver tissue samples with 27K DNA beadchips, one CpG site of the VPREB1 gene in non-tumor tissue showed significantly different methylation levels between alcohol users and non-users. Similarly, in HCC tumor tissues, methylation levels of CRISPLD1, PCDHB2, PCSK1, LXH1, KCTD8, TSHD3, and CXCL12 could distinguish alcohol users from non-users (36). In another study from Shen et al using 450K beadchips to analyze adjacent non-tumor tissues, 228 CpG sites were found to be significantly associated with HCV infection, and 17,207 CpG sites were associated with cirrhosis. Methylation levels on one CpG site of the OAZ3(MRPL9) gene exhibited significant difference between smokers and non-smokers. In the future, abnormal DNA methylation profiling maybe used to further distinguish HCC induced by different etiological and risk factors in HCC (e.g., HCV) (13).
Using 450K beadchips, Song also measured the DNA methylation profiles of 27 HCC samples and 20 para-tumor liver tissues. This study analyzed 10,775 differentially methylated sites distributed in or around gene promoter regions, 4% of which are known methylation sites. An additional 10,325 sites (covering 4,106 genes) may be differentially methylated in HCC (105). These DNA methylation sites are distributed abundantly in the CpG islands of promoter regions, which play important roles in regulatory signaling networks involving cell growth, gene expression regulation, apoptosis, and carcinogenesis. CpG sites of promoter regions of BMP4, CDKN2A, GSTP1 and NFATC1 genes showed the highest methylation levels. Similar studies have suggested that hypermethylation of promoter regions of tumor associated genes (i.e., suppressor genes and cell cycle genes) can inhibit their expression and thus increase the chance of HCC onset (141).
Many recent HCC-related epigenetic studies not only focused on the biological meaning of abnormal methylation, but also attempted to link biological discoveries with clinical applications. Samples taken from a cohort of 304 HCC patients were applied to a450K methylation beadchip and subjected to random forest processing method to obtain a diagnostic methylation signature composed of 36 methylation probes. This methylation signature, which is supported by data from HCV-associated HCC and alcohol-associated HCC, accurately predicted survival rates independent of clinical and pathological risk factors (e.g., multiple hepatic nodes, platelet count), and independently predicted the HCC recurrence rate. In addition, patients displaying a particular methylation signature and mRNA signature were more inclined to express the molecular subclass of proliferation with progenitor cell features (37,38). Zheng et al. analyzed global DNA sequencing data from 800 HCC samples, selected ten sites out of 80 hypermethylated promoter sites as candidate biomarkers to distinguish between tumor and non-tumor tissues, and reported that the sensitivity and specificity were both close to 100%. The selected sites included the CDKL2, TBX15, and NKX6-2 genes which have been previously reported. By integrative analysis of DNA methylation and gene expression data, Zheng also identified 222 candidate epigenetic drivers involved in inflammatory response and metabolic pathways, several of which—SFN, SPP1, and TKT were significantly associated with overall survival rates of patients (35).
Advances in next generation sequencing based examination techniques
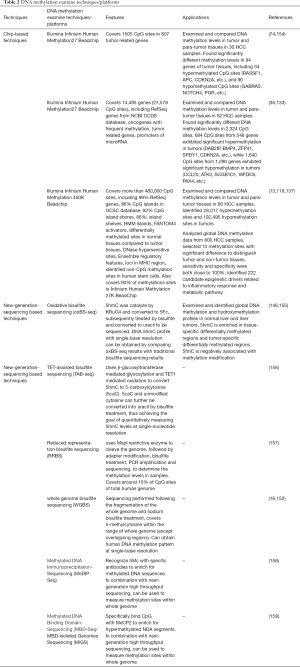
As of now, bisulfite sequencing remains the gold standard of DNA methylation profiling analysis (142). Recently, however, new examination techniques based on next generation sequencing have been developed and utilized in DNA methylation study. When using sequencing techniques to acquire methylation data, designing probes for known methylation levels is unnecessary; thus there are no constraints from previous experiments. However, the replicability of sequencing techniques is incomparable to that of chip array, and the processing of sequencing data is much more complicated. Sequencing based methylation analysis needs to be improved and optimized before it is fit for clinical application.
It has recently been discovered that the formation of 5-methylcytosine (from methylation of cytosine) can be modified by ten-eleven translocation methylcytosine dioxygenase, which oxidizes 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (143,144). Hydroxymethylation, as one of several DNA demethylation mechanisms, was found to be related to gene expression regulation and tumorigenesis (88,145). Traditional bisulfite sequencing can scarcely distinguish 5hmC from5-Mc, thus restricting the study of hydroxymethylation to some extent. Oxidative bisulfite sequencing (oxBS-seq) is the first method to quantitatively measure 5hmc levels in genome DNA at single-base resolution, and is distinguished by its simplicity, efficiency, and simultaneous collection both methylation and demethylation data (146). The oxBS-seq technique uses highly selective oxidative potassium perruthenate to convert 5-hmc into 5-formylcytosine (5fC), which is then converted into uracil by bisulfite treatment before finally being sequenced. Genome DNA 5hmc patterns can be obtained by comparing and mapping 5fC sequencing results with traditional bisulfite sequencing on the same sample (146). Meanwhile, another new sequencing technique, TET-assisted bisulfite sequencing (TAB-seq), uses β-glucosyltransferase mediated glycosylation and TET1 mediated oxidation to convert 5hmC to 5-carboxylcytosine (5caC). 5caC and unmodified cytosine can further be converted into uracil by bisulfite treatment, thus achieving the goal of quantitatively measuring 5hmC levels at single-nucleotide resolution (147).
Li et al., using the oxBS-seq technique, identified the global DNA methylation and hydroxymethylation signatures of normal livers and liver tumors at single-base resolution. 5hmC was enriched in tissue-specific DMRs and tumor-specific DMRs, and was inversely correlated with methylation modification (148). Expanding the findings from previous experiments showing that 5hmC is abundant in gene promoter regions, gene coding regions, and distal cis-regulatory elements-like enhancers (144), recent studies have reported that 5hmC is also enriched in CpG island shores (148).
Another widely used technique based on bisulfite sequencing is reduced representation bisulfite sequencing (RRBS). It uses MspI restrictive enzyme to cleave the genome, followed by adapter modification, bisulfite treatment, PCR amplification and sequencing, to determine the methylation levels in samples (149). RRBS covers most of CpG island and promoter sites, but this coverage only accounts for 10% of all CpG sites of human genome, making it difficult to monitor the genomic region’s lack of CpG sites (150).
Whole genome bisulfite sequencing (WGBS) is another technique based on bisulfite sequencing. The sequencing is performed following the fragmentation of the whole genome and sodium bisulfite treatment, which covers 5-methylcytosine within the range of whole genome (except overlapping regions). The WGBS method resolves the issues of low resolution, biased sequencing specificity, and partial genome analysis that are characteristic of traditional techniques (142,151,152). Lister et al. performed WGBS and obtained human methylation signatures at single-base resolution (153), but this technique has not yet been used for analyzing clinical samples (151). Considering the vast amount of data generated by WGBS, appropriate data processing approaches remain to be explored, and consistency between experimental and clinical results needs to be further verified (see Table 2).
Full table
Methylated DNA immunoprecipitation sequencing (mDIP-seq) and methylated DNA binding domain sequencing (MBD-seq) are global methylation analysis technique based on DNA methylation enrichment (142). In previous studies MeDIP-chip and MBD-chip have been widely used (156,160). In combination with next generation sequencing, MeDIP-Seq and MBD-Seq can also be suitable technique based on DNA methylation enrichment (142). In previous studies MeDIP-chip and MBD-chip have been widely used (156,160). In combination with next generation sequencing, MeDIP-Seq and MBD-Seq can also be suitable for global DNA methylation analysis. Down et al. obtained the methylome data from mammals using the MeDIP technique (158). Serre et al. examined DNA methylation in three homogenous tumor cell lines with MBD-seq, and reported methylation regions consistent with previous results as well as hundreds of new methylation regions, indicating the accuracy and sensitivity of this technique (159).
Prospective goals and future plan
Infinium Methylation EPIC Kit
Improvement in DNA methylation research in HCC relies on the advancement of methylation sequencing techniques. Following the gradual acceptance and usage of the 450k DNA methylation array, a new generation of DNA methylation array, the Illumina Infinium Methylation EPIC BeadChip, has been developed. As confirmed by the ENCODE and FANOM5 projects, this EPICchip can measure the methylation data of around 850,000 CpG sites from whole human genome, which covers >90% sites of 450K beadchip, in addition with 333,265 CpG sites in enhancer regions (155,157,161,162). A better genome wide association study can be performed with theEPIC chip owing to its sufficient probe coverage of CpG islands, gene coding regions, and promoters and enhancers, which provide an advanced technical platform for HCC epigenetic research.
scTrio-seq
Current single-cell methylation sequencing techniques include single-cell reduced-representation bisulfite sequencing (scRRBS) (163) and single-cell bisulfite sequencing (scBS) (164). scRRBS has been used in dynamic pattern analysis of DNA methylation during early embryo development (161). The recently introduced single-cell triple omics sequencing (scTrio-seq), first described by Hou et al., can simultaneously analyze the genomic copy number variation (CNV), DNA methylome, and transcriptome in a single mammalian cell, and thus can differentiate the contribution of genomic and epigenomic heterogeneities to the transcriptomic heterogeneity. In Hou’s work, the single-cell CNV, RNA expression, and methylation levels measure by scTrio-seq were proved to be consistent with results from scRRBS. Interestingly, though large-scale CNV caused proportional changes of RNA expression levels, the DNA methylation levels were rarely affected (165).
As elucidated by multiple previous studies, gene expression levels are negatively correlated with DNA methylation of gene promoter regions, but positively correlated with DNA methylation of gene bodies (166). Similar results were confirmed by Hou et al.: overall hypomethylation in HCC cells, negative correlation between promoter methylation level and RNA expression, and positive correlation between gene body methylation and RNA expression (165). Moreover, HCC cells were categorized into two subpopulations using scTrio-seq and unsupervised hierarchical clustering analysis. One subpopulation only accounts for a small portion of a given tumor, but may express high levels of cell invasion biomarkers. These subpopulations tend to escape immune surveillance, making them difficult to detect when systemically monitoring for tumors. Filtering and capturing different subpopulations of tumor cells will be an important topic for future cancer research (165).
Cell-free DNA from plasma
The majority of circulating cell-free DNA (ccfDNA) originates from apoptotic cells, and very few are from nucleated blood cells. It is therefore reasonable to study circulating tumor DNA via examining plasma (167,168). Several recent studies have explored the potential focusing cell-free nucleic acid in plasma as a biomarker to screen for tumors and evaluate prognoses and therapeutic effects (87,169-171). The use of whole genome methylation analysis to measure methylation of ccfDNA has begun, and may prove to be a novel way for the early detection of HCC (164,172). However, ccfDNA is usually highly fragmented, and its levels are affected by both tumor and non-tumor tissues (167). Moreover, during the early phases of cancer, non-tumor liver tissues may release more ccfDNA than tumor tissues. Therefore, the widespread usage of ccfDNA methylation analysis is greatly limited at this time (102,173).
MCTA-seq developed by Wen et al. achieved the goal of examining genome-scale hypermethylated CpG island in ccfDNA of HCC cells. The target sequence detection rate for the human white blood cell genome can be 93.3% using MCTA-seq analysis. Equipped with MCTA-seq, Wen et al. analyzed tissue and plasma sample from 151 HCC patients, and found four biomarkers with high specificity—RGS10, ST8SIA6, RUNX2, VIM, the methylation levels of which were positively correlated with the tumor size. Another type of biomarker revealed by MCTA-seq has higher sensitivity than ALT in detecting HCC-caused hepatocyte death. Joint application of these two biomarkers can be used to distinguish samples from HCC patients, cirrhosis patients, or healthy individuals, further improving the diagnosis rate of HCC when combined with current diagnosis criteria (i.e., AFP) (102).
As a non-invasive tumor test, hypermethylated CpG island analysis of ccfDNA has been drawing increasing attention. The hypermethylation of CDKL2, CDKN2A, HIST1H3G, STEAP4, and ZNF154 in ccfDNA can be detected in 37–63% of HCC patients (36). Based on several previous studies, gene methylation levels of CDKN2A/INK4(p16), RASSF1A, GSTP1, and MGMT in HCC cells were 30–70%, 85%, 50–90%, and 40%, respectively (87,92,96,106-108,174-176). Another study recruiting 25,000 participants found that the methylation levels of RASSF1A, CDKN2A, and INK4B (p15) in ccfDNA could predict the progression of HCC in later stages (161). The feasibility of examining DNA methylation in ccfDNA was confirmed by this study, and similar conclusions were also reached in other studies on solid tumors, although a larger-scale cohort study on HCC remains warranted (177-179).
In summary, despite a number of current genome-scale epigenetics studies in HCC, methylation data analysis is limited by various factors: huge gaps between the biological meaning and clinical meaning of the data, high heterogeneity of tumor tissues, and changeable cellular components. Therefore, future whole genome studies should include laser microdissection and high-speed flow cytometry cell sorting to isolate tumor cells from adjacent cells (17,180,181).
Furthermore, the correlation between abnormal DNA methylation and consequent dysfunction should be strictly evaluated. Even though an abundance of data may suggest the important role of candidate DNA methylation or candidate tumor suppressor genes in the pathogenesis and progression of HCC, further evidence should be obtained by manipulating in vivo DNA methylation of specific sites is required, which can more directly disclose the relationship between abnormal methylation and altered gene expression.
Last but not least, much of the current research was focused on or limited on the levels of cell or molecular biology, lacking any connection to existing clinical data, especially DNA methylation marker and clinical application. Therefore, future preclinical studies of tissues, cell, and molecules require more evidence from clinical samples and in vivo experiments. At the same time, the potential biomarkers of abnormal DNA methylation raised from HCC cells should be gradually introduced into clinical application for the purpose of evaluating cancer progression, predicting prognosis, monitoring recurrence, and early diagnosis.
Acknowledgments
We would like to thank Professor Jin Gu of Tsinghua University for his constructive suggestion.
Funding: This work was supported by Beijing Municipal Science& Technology Commission research fund (No. Z171100000417004).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- El-Serag HB. Hepatocellular Carcinoma. N Engl J Med 2011;365:1118-27. [Crossref] [PubMed]
- Harada K, Baba Y, Ishimoto T, et al. LINE-1 Methylation Level and Patient Prognosis in a Database of 208 Hepatocellular Carcinomas. Ann Surg Oncol 2015;22:1280-7. [Crossref] [PubMed]
- Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 2014;63:844-55. [Crossref] [PubMed]
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. [Crossref] [PubMed]
- Chen W, Zheng R, Baade DP, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Xiang W, Shi J, Li P, et al. Estimation of cancer cases and deaths attributable to infection in China. Cancer Causes Control 2011;22:1153-61. [Crossref] [PubMed]
- Li RC, Yang JY, Gong J, et al. Efficacy of hepatitis B vaccination on hepatitis B prevention and on hepatocellular carcinoma. Zhonghua Liu Xing Bing Xue Za Zhi 2004;25:385-7. [PubMed]
- Chang MH, Chen CJ, Lai MS, et al. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. N Engl J Med 1997;336:1855-9. [Crossref] [PubMed]
- Yu SZ, Chen G, Zhi X, et al. Primary liver cancer: natural toxins and prevention in China. J Toxicol Sci 1998;23:143-7. [Crossref] [PubMed]
- Liang X, Bi S, Yang W, et al. Epidemiological setosurvey of hepatitis B in China—declining HBV prevalence due to hepatitis B vaccination. Vaccine 2009;27:6550-7. [Crossref] [PubMed]
- Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol 2006;21:15-21. [Crossref] [PubMed]
- Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Current genomics 2011;12:130-7. [Crossref] [PubMed]
- Shen J, Wang S, Zhang Y, et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics 2013;8:34-43. [Crossref] [PubMed]
- Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-2. [Crossref] [PubMed]
- Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol 2010;16:418-24. [Crossref] [PubMed]
- Li X, Wang Y, Hu J, et al. Research progress on epigenetic of liver cancer. J Mod Oncol 2016;24:1501-4.
- Hardy T, Mann DA. Epigenetics in liver disease: from biology to therapeutics. Gut 2016;65:1895-905. [Crossref] [PubMed]
- Waddington CH. The epigenotype. 1942. Int J Epidemiol 2012;41:10-3. [Crossref] [PubMed]
- Rountree MR, Bachman KE, Herman JG, et al. DNA methylation, chromatin inheritance, and cancer. Oncogene 2001;20:3156-65. [Crossref] [PubMed]
- Stefanska B, Huang J, Bhattacharyya B, et al. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res 2011;71:5891-903. [Crossref] [PubMed]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Medicine 2011;17:330-9. [Crossref] [PubMed]
- Gal-Yam EN, Saito Y, Egger G, et al. Cancer Epigenetics: Modifications, Screening, and Therapy. Annu Rev Med 2008;59:267-80. [Crossref] [PubMed]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12-27. [Crossref] [PubMed]
- Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene 2010;29:2309-24. [Crossref] [PubMed]
- Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 2013;13:123-35. [Crossref] [PubMed]
- Yu J, Ni M, Xu J, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. Bmc Cancer 2002;2:29. [Crossref] [PubMed]
- Toraño EG, Petrus S, Fernandez AF, et al. Global DNA hypomethylation in cancer: review of validated methods and clinical significance. Clin Chem Lab Med 2012;50:1733-42. [Crossref] [PubMed]
- Hernandez-Vargas H, Lambert MP, Le Calvez-Kelm F, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PloS One 2010;5:e9749 [Crossref] [PubMed]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012;13:679-92. [Crossref] [PubMed]
- Yang X, Gao L, Zhang S. Comparative pan-cancer DNA methylation analysis reveals cancer common and specific patterns. Brief Bioinform 2017;18:761-73. [PubMed]
- Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317-25. [Crossref] [PubMed]
- BLUEPRINT consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol 2016;34:726-37. [Crossref] [PubMed]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer-a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107-16. [Crossref] [PubMed]
- De Carvalho DD, Sharma S, You JS, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 2012;21:655-67. [Crossref] [PubMed]
- Zheng Y, Huang Q, Ding Z, et al. Genome-wide DNA methylation analysis identifies candidate epigenetic markers and drivers of hepatocellular carcinoma. Brief Bioinform 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology 2012;55:1799-808. [Crossref] [PubMed]
- Villanueva A, Portela A, Sayols S, et al. DNA methylation− based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015;61:1945-56. [Crossref] [PubMed]
- Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009;69:7385-92. [Crossref] [PubMed]
- Lee S, Lee HJ, Kim JH, et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol 2003;163:1371-8. [Crossref] [PubMed]
- Novik KL, Nimmrich I, Genc B, et al. Epigenomics: genome-wide study of methylation phenomena. Curr Issues Mol Biol 2002;4:111-28. [PubMed]
- Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology 2003;4:233-50. [Crossref] [PubMed]
- Kuo KC, McCune RA, Gehrke CW, et al. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res 1980;8:4763-76. [Crossref] [PubMed]
- Fraga MF, Uriol E, Borja DL, et al. High-performance capillary electrophoretic method for the quantification of 5-methyl 2'-deoxycytidine in genomic DNA: application to plant, animal and human cancer tissues. Electrophoresis 2002;23:1677-81. [Crossref] [PubMed]
- Schmitt F, Oakeley EJ, Jost JP. Antibiotics induce genome-wide hypermethylation in cultured Nicotiana tabacum plants. J Biol Chem 1997;272:1534-40. [Crossref] [PubMed]
- Wu J, Issa JP, Herman J, et al. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci U S A 1993;90:8891-5. [Crossref] [PubMed]
- Oakeley EJ, Schmitt F, Jost JP. Quantification of 5-methylcytosine in DNA by the chloroacetaldehyde reaction. Biotechniques 1999;27:744-6, 748-50, 752. [PubMed]
- Oakeley EJ, Podesta A, Jost JP. Developmental changes in DNA methylation of the two tobacco pollen nuclei during maturation. Proc Natl Acad Sci U S A 1997;94:11721-5. [Crossref] [PubMed]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 1975;98:503-17. [Crossref] [PubMed]
- Frommer M, Mcdonald LE, Millar DS, et al. A Genomic Sequencing Protocol That Yields A Positive Display of 5-Methylcytosine Residues in Individual DNA Strands. Proc Natl Acad Sci U S A 1992;89:1827-31. [Crossref] [PubMed]
- Melnikov AA, Gartenhaus RB, Levenson A, et al. MSRE-PCR for analysis of gene-specific DNA methylation. Nucleic Acids Res 2005;33:e93 [Crossref] [PubMed]
- Herman JG, Graff JR, Myohanen S, et al. Methylation-Specific PCR: A Novel PCR Assay for Methylation Status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821-6. [Crossref] [PubMed]
- Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res 1997;25:2529-31. [Crossref] [PubMed]
- Campan M, Weisenberger JD, Trinh B, et al. MethyLight. Humana Press, 2009:325-37.
- Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32 [Crossref] [PubMed]
- Maekawa M, Sugano K, Kashiwabara H, et al. DNA methylation analysis using bisulfite treatment and PCR-single-strand conformation polymorphism in colorectal cancer showing microsatellite instability. Biochem Biophys Res Commun 1999;262:671-6. [Crossref] [PubMed]
- Burri N, Chaubert P. Complex methylation patterns analyzed by single-strand conformation polymorphism. Biotechniques 1999;26:232-4. [PubMed]
- Bianco T, Hussey D, Dobrovic A. Methylation-sensitive, single-strand conformation analysis (MS-SSCA): A rapid method to screen for and analyze methylation. Human Mutation 1999;14:289-93. [Crossref] [PubMed]
- Aggerholm A, Guldberg P, Hokland M, et al. Extensive intra- and interindividual heterogeneity of p15INK4B methylation in acute myeloid leukemia. Cancer Res 1999;59:436-41. [PubMed]
- Worm J, Aggerholm A, Guldberg P. In-tube DNA methylation profiling by fluorescence melting curve analysis. Clin Chem 2001;47:1183-9. [PubMed]
- Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: A review. Human Mutation 2001;17:439-74. [Crossref] [PubMed]
- Gitan RS, Shi H, Chen CM, et al. Methylation-Specific Oligonucleotide Microarray: A New Potential for High-Throughput Methylation Analysis. Genome Res 2002;12:158-64. [Crossref] [PubMed]
- Hatada I, Hayashizaki Y, Hirotsune S, et al. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci U S A 1991;88:9523-7. [Crossref] [PubMed]
- Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res 1999;59:2307-12. [PubMed]
- Lisitsyn N, Wigler M. Cloning the difference between two complex genomes. Science 1993;259:946-51. [Crossref] [PubMed]
- Gonzalgo ML, Liang G, Spruck CH, et al. Identification and Characterization of Differentially Methylated Regions of Genomic DNA by Methylation-sensitive Arbitrarily Primed PCR. Cancer Res 1997;57:594-9. [PubMed]
- Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Human Molecular Genetics 1999;8:459-70. [Crossref] [PubMed]
- Yan PS, Chen CM, Shi H, et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res 2001;61:8375-80. [PubMed]
- Nishida N, Kudo M, Nagasaka T, et al. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology 2012;56:994-1003. [Crossref] [PubMed]
- Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst 2002;94:755-61. [Crossref] [PubMed]
- Nagasaka T, Koi M, Kloor M, et al. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology 2008;134:1950-60. [Crossref] [PubMed]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 1997;25:2532-4. [Crossref] [PubMed]
- Umer M, Qureshi SA, Hashmi ZY, et al. Promoter hypermethylation of Wnt pathway inhibitors in hepatitis C virus - induced multistep hepatocarcinogenesis. Virol J 2014;11:117. [Crossref] [PubMed]
- Ushijima T, Okochi-Takada E. Aberrant methylations in cancer cells: Where do they come from? Cancer Sci 2005;96:206-11. [Crossref] [PubMed]
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002;31:339-46. [Crossref] [PubMed]
- Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006;130:1117-28. [Crossref] [PubMed]
- Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest 2007;117:2713-22. [Crossref] [PubMed]
- Kranenburg O, Gebbink MFBG, Voest EE. Stimulation of angiogenesis by Ras proteins. Biochimica Et Biophysica Acta 2004;1654:23-37. [PubMed]
- Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell 2004;6:447-58. [Crossref] [PubMed]
- Ammerpohl O, Pratschke J, Schafmayer C, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer 2012;130:1319-28. [Crossref] [PubMed]
- Lehmann U, Berg-Ribbe I, Wingen LU, et al. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin Cancer Res 2005;11:3654-60. [Crossref] [PubMed]
- Csepregi A, Rocken C, Hoffmann J, et al. APC promoter methylation and protein expression in hepatocellular carcinoma. J Cancer Res Clin Oncol 2008;134:579-89. [Crossref] [PubMed]
- Moribe T, Iizuka N, Miura T, et al. Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. Int J Cancer 2009;125:388-97. [Crossref] [PubMed]
- Hua D, Hu Y, Wu YY, et al. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol 2011;91:455-60. [Crossref] [PubMed]
- Liu JB, Zhang YX, Zhou SH, et al. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol 2011;17:4718-24. [Crossref] [PubMed]
- Cho S, Lee JH, Cho SB, et al. Epigenetic methylation and expression of caspase 8 and survivin in hepatocellular carcinoma. Pathol Int 2010;60:203-11. [Crossref] [PubMed]
- McConnell BB, Vertino PM. TMS1/ASC: the cancer connection. Apoptosis 2004;9:5-18. [Crossref] [PubMed]
- Yeo W, Wong N, Wong WL, et al. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int 2005;25:266-72. [Crossref] [PubMed]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 2013;6:10. [Crossref] [PubMed]
- Wong CM, Lee JM, Ching YP, et al. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res 2003;63:7646-51. [PubMed]
- Shin SH, Kim BH, Jang JJ, et al. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci 2010;25:1152-9. [Crossref] [PubMed]
- Tischoff I. DNA methylation in hepatocellular carcinoma. World J Gastroenterol 2008;14:1741-8. [Crossref] [PubMed]
- Yang B, Guo M, Herman JG, et al. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol 2003;163:1101-7. [Crossref] [PubMed]
- Li Binkui, Liu W, Wang L, et al. CpG island methylator phenotype associated with tumor recurrence in tumor-node-metastasis stage I hepatocellular carcinoma. Ann Surg Oncol 2010;17:1917-26. [Crossref] [PubMed]
- Kaneto H, Sasaki S, Yamamoto H, et al. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut 2001;48:372-7. [Crossref] [PubMed]
- Lü GL, Wen JM, Xu JM, et al. Relationship between TIMP-3 expression and promoter methylation of TIMP-3 gene in hepatocellular carcinoma. Zhonghua Bing Li Xue Za Zhi 2003;32:230-3. [PubMed]
- Zhang YJ, Ahsan H, Chen Y, et al. High frequency of promoter hypermethylation of RASSF1A and p16 and its relationship to aflatoxin B1-DNA adduct levels in human hepatocellular carcinoma. Mol Carcinog 2002;35:85-92. [Crossref] [PubMed]
- Takagi H, Sasaki S, Suzuki H, et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol 2008;43:378-89. [Crossref] [PubMed]
- Revill K, Wang T, Lachenmayer A, et al. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 2013;145:1424-35.e1-25.
- Yoshikawa H, Matsubara K, Qian GS, et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet 2001;28:29-35. [Crossref] [PubMed]
- Um TH, Kim H, Oh BK, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol 2011;54:939-47. [Crossref] [PubMed]
- Stefanska B, Suderman M, Machnes Z, et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin Cancer Res 2014;20:3118-32. [Crossref] [PubMed]
- Wen L, Li J, Guo H, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015;25:1250-64. [Crossref] [PubMed]
- Lai HJ, Lo SJ. Epigenetic methylation of TIMP-3 may play a role in HBV-associated hepatocellular carcinoma. Chang Gung Med J 2005;28:453-5. [PubMed]
- Harder J, Opitz OG, Brabender J, et al. Quantitative promoter methylation analysis of hepatocellular carcinoma, cirrhotic and normal liver. Int J Cancer 2008;122:2800-4. [Crossref] [PubMed]
- Song MA, Tiirikainen M, Kwee S, et al. Elucidating the Landscape of Aberrant DNA Methylation in Hepatocellular Carcinoma. PloS One 2013;8:e55761 [Crossref] [PubMed]
- Zhang YJ, Chen Y, Ahsan H, et al. Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma. Cancer Letters 2005;221:135-43. [Crossref] [PubMed]
- Zhong S, Tang MW, Yeo W, et al. Silencing of GSTP1 Gene by CpG Island DNA Hypermethylation in HBV-associated Hepatocellular Carcinomas. Clin Cancer Res 2002;8:1087-92. [PubMed]
- Zhang YJ, Chen Y, Ahsan H, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutation in hepatocellular carcinoma. Int J Cancer 2003;103:440-4. [Crossref] [PubMed]
- Mannello F, Gazzanelli G. Tissue inhibitors of metalloproteinases and programmed cell death: Conundrums, controversies and potential implications. Apoptosis 2001;6:479-82. [Crossref] [PubMed]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. [Crossref] [PubMed]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009;10:691-703. [Crossref] [PubMed]
- Ogino S, Nosho K, Kirkner GJ, et al. A Cohort Study of Tumoral LINE-1 Hypomethylation and Prognosis in Colon Cancer. J Natl Cancer Inst 2008;100:1734-8. [Crossref] [PubMed]
- Iwagami S, Baba Y, Watanabe M, et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg 2013;257:449-55. [Crossref] [PubMed]
- Pattamadilok J, Huapai N, Rattanatanyong P, et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer 2008;18:711-7. [Crossref] [PubMed]
- Shigaki H, Baba Y, Watanabe M, et al. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer 2013;16:480-7. [Crossref] [PubMed]
- Irahara N, Nosho K, Baba Y, et al. Precision of Pyrosequencing Assay to Measure LINE-1 Methylation in Colon Cancer, Normal Colonic Mucosa, and Peripheral Blood Cells. J Mol Diagn 2010;12:177-83. [Crossref] [PubMed]
- Ogino S, Nosho K. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer 2008;122:2767-73. [Crossref] [PubMed]
- Yang AS, Estécio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004;32:e38 [Crossref] [PubMed]
- Baylin SB. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin Cancer Biol 2002;12:331-7. [Crossref] [PubMed]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92. [Crossref] [PubMed]
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta 2007;1775:138-62.
- Esteller M. DNA methylation and cancer therapy: new developments and expectations. Curr Opin Oncol 2005;17:55-60. [Crossref] [PubMed]
- Chen RZ, Pettersson U, Beard C, et al. DNA hypomethylation leads to elevated mutation rates. Nature 1998;395:89-93. [Crossref] [PubMed]
- Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science 2003;300:489-92. [Crossref] [PubMed]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042-54. [Crossref] [PubMed]
- Kim M, Trinh BN, Long TI, et al. Laird, Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res 2004;32:5742-9. [Crossref] [PubMed]
- Kondo Y, Shen L, Suzuki S, et al. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol Res 2007;37:974-83. [Crossref] [PubMed]
- Nishida N, Nagasaka T, Nishimura T, et al. Aberrant Methylation of Multiple Tumor Suppressor Genes in Aging Liver, Chronic Hepatitis, and Hepatocellular Carcinoma. Hepatology 2008;47:908-18. [Crossref] [PubMed]
- Nomoto S, Kinoshita T, Kato K, et al. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer 2007;97:1260-5. [Crossref] [PubMed]
- Su H, Zhao J, Xiong Y, et al. Large-scale analysis of the genetic and epigenetic alterations in hepatocellular carcinoma from Southeast China. Mutat Res 2008;641:27-35. [Crossref] [PubMed]
- Su PF, Lee TC, Lin PJ, et al. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int J Cancer 2007;121:1257-64. [Crossref] [PubMed]
- Zhang C, Guo X, Jiang G, et al. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer 2008;123:998-1004. [Crossref] [PubMed]
- Weisenberger D, Van Den Berg D, Pan F, et al. Comprehensive DNA methylation analysis on the Illumina Infinium assay platform. Illumina, San Diego, 2008.
- Kim YH, Lee HC, Kim SY, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol 2011;18:2338-47. [Crossref] [PubMed]
- Rajendram R, Ferreira JC, Grafodatskaya D, et al. Assessment of methylation level prediction accuracy in methyl-DNA immunoprecipitation and sodium bisulfite based microarray platforms. Epigenetics 2011;6:410-5. [Crossref] [PubMed]
- Yuen RK, Peñaherrera MS, von Dadelszen P, et al. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet 2010;18:1006-12. [Crossref] [PubMed]
- Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288-95. [Crossref] [PubMed]
- Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 2009;41:1350-3. [Crossref] [PubMed]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178-86. [Crossref] [PubMed]
- Ogoshi K, Hashimoto S, Nakatani Y, et al. Genome-wide profiling of DNA methylation in human cancer cells. Genomics 2011;98:280-7. [Crossref] [PubMed]
- Tischoff I, Tannapfel A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol 2008;14:1741-8. [Crossref] [PubMed]
- Li N, Ye M, Li Y, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 2010;52:203-12. [Crossref] [PubMed]
- Laird A, Thomson JP, Harrison DJ, et al. 5-hydroxymethylcytosine profiling as an indicator of cellular state. Epigenomics 2013;5:655-69. [Crossref] [PubMed]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014;156:45-68. [Crossref] [PubMed]
- Kudo Y, Tateishi K, Yamamoto K, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci 2012;103:670-6. [Crossref] [PubMed]
- Booth MJ, Branco MR, Ficz G, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012;336:934-7. [Crossref] [PubMed]
- Yu M, Hon GC, Szulwach KE. Base-Resolution Analysis of 5-Hydroxymethylcytosine in the Mammalian Genome. Cell 2012;149:1368-80. [Crossref] [PubMed]
- Li X, Liu Y, Salz T, et al. Whole-genome analysis of the methylome and hydroxymethylome in normal and malignant lung and liver. Genome Res 2016;26:1730-41. [Crossref] [PubMed]
- Meissner A, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 2005;33:5868-77. [Crossref] [PubMed]
- Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature 2014;511:606-10. [Crossref] [PubMed]
- Jeong HM, Lee S, Chae H, et al. Efficiency of methylated DNA immunoprecipitation bisulphite sequencing for whole-genome DNA methylation analysis. Epigenomics 2016;8:1061-77. [Crossref] [PubMed]
- Lister R, O'Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008;133:523-36. [Crossref] [PubMed]
- Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315-22. [Crossref] [PubMed]
- Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res 2006;16:383-93. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 2005;37:853-62. [Crossref] [PubMed]
- Lizio M, Harshbarger J, Shimoji H, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015;16:22. [Crossref] [PubMed]
- Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 2008;26:779-85. [Crossref] [PubMed]
- Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 2010;38:391-9. [Crossref] [PubMed]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 2006;126:1189-201. [Crossref] [PubMed]
- Siggens L, Ekwall K. Epigenetics, chromatin and genome organization: recent advances from the ENCODE project. J Intern Med 2014;276:201-14. [Crossref] [PubMed]
- Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011;6:692-702. [Crossref] [PubMed]
- Guo H, Zhu P, Wu X, et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res 2013;23:2126-35. [Crossref] [PubMed]
- Smallwood SA, Lee HJ, Angermueller C, et al. Single-Cell Genome-Wide Bisulfite Sequencing for Assessing Epigenetic Heterogeneity. Nat Methods 2014;11:817-20. [Crossref] [PubMed]
- Hou Y, Guo H, Cao C, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 2016;26:304-19. [Crossref] [PubMed]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet 2013;14:204-20. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. The Origin and Mechanism of Circulating DNA. Ann N Y Acad Sci 2000;906:161-8. [Crossref] [PubMed]
- Wong IH, Lo YM, Yeo W, et al. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res 2000;6:3516-21. [PubMed]
- Wong IH, Lo YM, Zhang J, et al. Detection of Aberrant p16 Methylation in the Plasma and Serum of Liver Cancer Patients. Cancer Res 1999;59:71-3. [PubMed]
- Zhang YJ, Wu HC, Shen J, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res 2007;13:2378-84. [Crossref] [PubMed]
- Sun K, Jiang P, Chan KC, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015;112:E5503-12. [Crossref] [PubMed]
- Gu H, Smith ZD, Bock C, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 2011;6:468-81. [Crossref] [PubMed]
- Hui AM, Sakamoto M, Kanai Y, et al. Inctivation of p16INK4 in hepatocellular carcinoma. Hepatology 1996;24:575-9. [Crossref] [PubMed]
- Matsuda Y, Ichida T, Matsuzawa J, et al. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology 1999;116:394-400. [Crossref] [PubMed]
- Zhang YJ, Rossner J Jr, Chen Y, et al. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int J Cancer 2006;119:985-91. [Crossref] [PubMed]
- Li X, Zhou F, Jiang C, et al. Identification of a DNA Methylome Profile of Esophageal Squamous Cell Carcinoma and Potential Plasma Epigenetic Biomarkers for Early Diagnosis. Plos One 2014;9:e103162 [Crossref] [PubMed]
- Sookoian S, Pirola CJ. Cell-free DNA methylation as liquid biopsy for the assessment of fibrosis in patients with nonalcoholic steatohepatitis: a gap between innovation and implementation. Hepatobiliary Surg Nutr 2017;6:117-21. [Crossref] [PubMed]
- Teufel U, Peccerella T, Engelmann G, et al. Detection of carcinogenic etheno-DNA adducts in children and adolescents with non-alcoholic steatohepatitis (NASH). Hepatobiliary Surg Nutr 2015;4:426-35. [PubMed]
- Linhart KB, Glassen K, Peccerella T, et al. The generation of carcinogenic etheno-DNA adducts in the liver of patients with nonalcoholic fatty liver disease. Hepatobiliary Surg Nutr 2015;4:117-23. [PubMed]
- Ghosh S, Banerjee S. Whole-genome mutational analysis: cause and effect of noncoding and structural mutations in liver cancer. Hepatobiliary Surg Nutr 2017;6:57-9. [Crossref] [PubMed]


