
Estrogen receptor β and estrogen receptor α36 predict differential outcome of patients with breast cancer
Introduction
Breast cancer is the most commonly diagnosed cancer in women worldwide (1). Over the last decades, many classic signaling molecules, such as estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2), have been identified and applied for breast cancer diagnosis and prognosis. However, the prognostic value of these traditional molecules for breast cancer still has certain limitations. Identification of novel markers is very important for improving prognostic judgment and clinical management.
ERα and ERβ are two members of the steroid nuclear receptor superfamily of ligand-activated transcription factors. Although they mediate estrogen signaling by binding to the estrogen-response element of target genes with 96% identity in DNA-binding domains, ERs exhibit different biological effects due to the considerable difference in ligand-binding domains (2). ERα is commonly considered as a tumor promoter and is involved in the carcinogenesis and progression of breast cancer. ERα66, the main type of ERα, serves as one of the most important markers for clinical molecular classification of human breast cancer. ERα36 is a novel variant of ERα that has a molecular weight of 36 kD and differs from the original 66 kD ERα (ERα66), lacking both transcriptional activation domains (AF-1 and AF-2) but retaining the DNA-binding domain and partial dimerization and ligand-binding domains (3). ERα36 is mainly expressed in the cytoplasm and the plasma membrane. Shi et al. reported that patients with both high expression of ERα66 and ERα36 are less likely to be benefited from treatment of tamoxifen (4), a selective estrogen receptor modulator (SERM) with mixed agonist/antagonist activities that has been used widely to treat ERα66-positive breast cancer (5). A recent study has also revealed that ERα36 could mediate membrane-initiated signaling to enhance the agonist activity of tamoxifen (6). Our recent work demonstrated that tamoxifen could directly bind and activate ERα36 to enhance the stemness and metastasis of breast cancer cells via transcriptional stimulation of aldehyde dehydrogenase 1A1 (ALDH1A1) (7). These findings suggest that ER-α36 may have predictive and prognostic value in breast cancer. ERβ, the second ER, has been discovered and studied for over 20 years, but its biological function as well as its role as a prognostic or predictive factor in human breast cancer remains confliction. There are three ERβ isoforms in breast cancers, referred to ERβ1, ERβ2 (also known as ERβcx), and ERβ5 (8). ERβ1 is the wild type and the only fully functional isoform of ERβ (9,10). Our previous work revealed that ERβ1 could inhibit breast cancer cell growth through upregulation of p21 gene expression by binding with ID1 (11). Therefore, the clinical relevance of ERβ and ERα36 expression in human breast cancer is required to be further investigated.
In this study, we examined the expression of ERβ and ERα36 in the breast cancer samples from 124 patients by using immunohistochemistry and analyzed their clinical relevance.
Methods
Patients and tissue specimens
The breast cancer tissue specimens were obtained from 124 patients who were diagnosed with primary breast cancer and underwent surgery between 2008 and 2009 at Southwest Hospital, Third Military Medical University in China. The cohort included all the patients who were invasive breast cancer cases and did not receive preoperative radiotherapy or chemotherapy. Multiple clinical and pathological characteristics were obtained from the medical records and the original pathology reports, including age, histological tumor type and grade, tumor size, lymph node status and immunohistochemical (IHC) expressions of ER, PR and HER2. Follow-up information was updated every 6 months by telephone interview until April 2015. This study was performed with the consent of patients and approved by the Ethics Committee of the Southwest Hospital (ChiCTR-DCC-11001733).
IHC staining
Formalin-fixed paraffin embedded breast cancer tissue specimens were sliced into 4 µm sections and mounted on silanized slides. After placed at 56 °C overnight, the slides were dewaxed in xylene for 20 min and rehydrated in graded alcohols. For antigen retrieval, slides were placed in EDTA antigen retrieval solution and boiled for 20 min. After naturally cooling to room temperature and washing with PBS, endogenous peroxidase was blocked by using a 3% solution of hydrogen peroxide for 30 min 37 °C in the dark. Then slides were washed with PBS and incubated with primary antibodies of monoclonal mouse anti-human ERβ1 antibody (ab187291, Abcam, dilution 1:200) and monoclonal mouse anti-human ERα36 antibody (7) (Shinogen, China, dilution 1:200) at 4 °C overnight, respectively. Then slides were incubated with secondary antibody (DAKO, Denmark, K5007) at 37 °C for 30 min in the dark. After antibody incubation, slides were developed with DAB chromogenic reagent and counterstained with haematoxylin. The negative controls were performed by PBS replacement of primary antibodies.
Evaluation of Immunostaining
ERβ and ERα36 immunostaining signals were evaluated independently by two pathologists in a blinded manner. Brown nuclear staining for ERβ and brown cytoplasmic and membranous staining for ERα36 were considered positive. The staining intensity of positive tumor cells was scored as 0 (no), 1 (weak), 2 (moderate) and 3 (strong). The percentage of positively stained tumor cells was scored with 5 scales: 0 (none); 1 (<10%); 2 (11–25%); 3 (26–50%); 4 (51–75%); and 5 (>75%). The final scores were the sum of the intensity and the percentage score. With X-tile analysis (12), the cut-off value was determined as score 3 both for ERβ and ERα36. Thus, the scores that ≤3 were defined as “negative” expression, otherwise they were defined as “positive” expression.
Statistical analysis
The correlation between ERβ and ERα36 was analyzed by using Spearman’s rank test. The relationship between ERβ/ERα36 expression and clinicopathologic parameters was analyzed by using a two-tailed Chi-square test. Survival curves were generated by using Kaplan-Meier method with log-rank test. Univariate or multivariate analysis of prognostic factors was tested by using Cox proportional hazard regression model. All statistical analyses were performed by using SPSS software system (version 18.0; SPSS). P<0.05 was considered to be statistically significant.
Results
The expression pattern of ERβ and ERα36 in human breast cancer
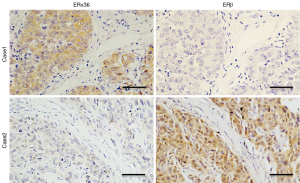
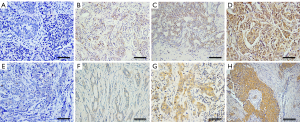
We measured the expression of ERβ and ERα36 in 124 human breast cancer tissues by IHC staining. Sixty-five cases (65/124, 52.4%, Table 1) showed positive expression of ERβ (ERβ+), with positive staining signals mainly in the nucleus (Figure S1A,B,C,D). Forty-nine cases (49/124, 39.5%, Table 1) were ERα36 positive (ERα36+), with predominately localization in the cytoplasm and cytomembrane (Figure S1E,F,G,H). There were 16 cases (16/124, 12.9%) showed positive and 26 cases (26/124, 21.0%) showed negative both for ERβ and ERα36. Spearman correlation analysis showed that ERβ expression was inversely correlated with ERα36 expression (r=−0.196, P=0.029, Figure 1). These results indicate that ERβ and ERα36 may have opposite clinical significance in human breast cancer.
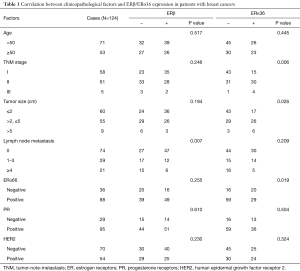
Full table
Association of ERβ/ERα36 expression with clinicopathological characteristics
To clarify the clinical significance of ERβ and ERα36 in human breast cancer, we compared ERβ/ERα36 expression with clinicopathological characteristics, including age, TNM stage, tumor size, lymph node metastasis, ERα66/PR/HER2 expression (Table 1). ERβ expression was inversely associated with lymph node metastasis (P=0.007). Whereas the expression of ERα36 was positively correlated to TNM stage (P=0.006), tumor size (P=0.026) and ERα66 expression (P=0.019). These results suggest that ERβ trends to be a protective factor, while ERα36 may act as a marker for aggressive human breast cancer.
Prognostic value of ERβ/ERα36 expression for patients with breast cancer
Kaplan-Meier analysis showed that the median disease-free survival (DFS) time of whole cohort were 68.0 (95% CI, 64.5–71.5) months. The patients with ERβ+ had significantly better DFS rate than those with ERβ-negative (ERβ−) (P=0.019, Figure 2A). Accordingly, the median DFS time [78.9 (95% CI, 75.6–82.1) months] in ERβ+ group was significantly longer than that in ERβ− group [64.6 (95% CI, 57.9–71.4) months]. Cox regression analysis was performed to determine the independence of several prognostic factors, including age, tumor size, lymph node metastasis, ERα66/PR/HER2 expression and ERβ/ERα36 expression. The result revealed that ERβ was an independent protective prognostic factor (HR=0.336, P=0.026, Table 2).

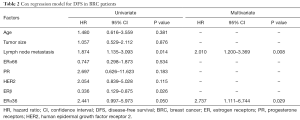
Full table
Opposite to the prognostic value of ERβ, patients with ERα36+ had poorer clinical outcome than those with ERα36- (P=0.043, Figure 2B). The 5-year DFS rates of patients with ERα36+ and ERα36- were 75.5% and 88.9%, respectively. Cox regression analysis revealed that ERα36 was an independent prognostic factor to predict poorer outcome of patients (HR=2.737, P=0.029, Table 2). Taken together, these results suggest that ERβ and ERα36 have contrarily prognostic role in human breast cancer.
Predictive model of combinative expression of ERβ and ERα36 in human breast cancer
To evaluate the predictive capability of combination of ERβ and ERα36 expression on the outcome of breast cancer patients, we divided patients into four groups: ERβ−/ERα36- group (N=26), ERβ+/ERα36- group (N=49), ERβ+/ERα36+ group (N=16) and ERβ−/ERα36+group (N=33). Kaplan-Meier analysis showed that ERβ−/ERα36+ group had the poorest DFS rate than other groups (P=0.014, Figure 2C). The DFS rate of ERβ+/ERα36+ group was similar to that of ERβ+/ERα36-group, whereas the DFS rates of the two groups were better than that of ERβ−/ERα36+ group. Moreover, in the patients with ERα36+, ERβ served as a potential independent protective prognostic marker (HR=0.144, P=0.064, Table 3). These results indicate that combination of ERβ and ERα36 expression may provide a better predictive model in human breast cancer.
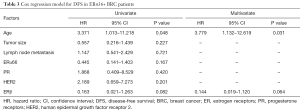
Full table
Discussion
Following the discovery of ERβ, investigators have sought to uncover its role in the progression and treatment of breast cancer. Many conflicting results have been reported. Chang et al. found that ERβ was highly expressed in tamoxifen-resistant breast cancer cells and appeared to indicate a poor response to endocrine treatment (13). Guo et al. reported that the tumor-free survival rate in patients with positive ERβ expression was significantly lower than that in patients with negative ERβ expression (14). Conversely, a lot of studies indicated that ERβ may act as a tumor suppressor and the presence of ERβ was associated with a favorable outcome (15-18). Thomas et al. found that ERβ inhibited both EMT and invasive capability of basal-like breast cancer cells either in vitro or in vivo (19,20). More recently, ERβ may target genes involved in the Wnt/β-catenin and the G1/S cell cycle phase checkpoint pathways to stimulate its growth-inhibitory effects in triple negative breast cancer cells (21). Cotrim et al. reported that ERβ inhibited breast cancer cell proliferation through inactivating of MAPK and PI3K signaling (22). Honma et al. indicated that ERβ was associated with a favorable survival for the patients after adjuvant tamoxifen monotherapy (23). Our results are consistent with that ERβ is a favorable prognostic factor. The level of ERβ is inversely correlated with lymph node metastasis. Moreover, the 5-year DFS rate is much higher in the patients with ERβ+ than those with ERβ−.
Approximately 70% of primary breast cancers express ERα66 (24), providing the rationale for the successful use of targeted endocrine therapies in breast cancer. However, not all patients with positive-ERα66 expression respond to endocrine treatment because of primary or secondary resistance. Recent studies have indicated that ERα36 is a very important factor in tamoxifen resistance. Shi et al. reported that patients with both high expressions of ERα66 and ERα36 are less likely to be benefited from tamoxifen treatment than those with high expression of ERα66 only (4). Structurally, ERα36 lacks the AF-1 and AF-2 domains, leading to the loss of intrinsic transcription activation. Identification of subcellular localization also revealed that ERα36 was mainly in the plasma membrane (~50%) and cytosol (~40%) (25). The special structure and distribution suggest that ERα36 may mainly act as membrane receptor to mediate rapid estrogen signaling (membrane-initiated signaling pathway). It has been demonstrated that ERα36 activated the MAPK/ERK and PI3K/AKT signaling pathways and promoted the tumor progression of breast cancer (6,26). Our results also revealed that ERα36 was a tumor promoter, which was associated with poorer clinicopathological characteristics, including the advanced TNM stages and the larger tumor size. Patients with ERα36+ also processed lower 5-year DFS rate than those with ERα36−.
Opposite to the function of ERα36, ERβ was found to be a suppressor of MAPK/ERK and PI3K/AKT signaling pathways (22). Lindberg et al. reported that ERβ could decrease pAKT level by up-regulating the expression of PTEN, thereby causing the impairment of proliferation ability and the enhancement of tamoxifen sensitivity for breast cancer cells (27). Wang et al. found that patients with ERβ (+)/pAKT(−) possessed longest survival time (28). Our work revealed that the patients of ERα36+/ERβ− had the poorer DFS rate than those of ERα36+/ERβ+. Moreover, ERβ could serve as an independent and favorable prognostic indicator in ERα36+ patients. Thus, the combinative expression of ERβ and ERα36 may provide a better prognosis model in human breast cancer.
In this study, the cut-off value of IHC scores for both ERβ and ERα36 was determined as 3 by X-tile analysis, which is computed by looking at the survival beforehand. Nevertheless, even if the same IHC scoring is applied, the cut-off value could not be directly applied in practice because of the small sample size of this study.
Conclusions
In conclusion, we demonstrate that both ERβ and ERα36 can act as prognostic indicators for human breast cancer. ERβ is a favorable indicator, while ERα36 is an unfavorable indicator. Combination of ERβ and ERα36 expression status has a better prognostic significance for breast cancer patients. However, the results need to be validated in further studies.
Acknowledgments
We would like to thank the Shinogen Company, Beijing, China for providing the primary ERα36 antibody for free, and especially Peipei Wang (Shinogen) for supporting our experiments.
Funding: This research was supported by National Science Foundation of China (No. 81372555).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was performed with the consent of patients and approved by the Ethics Committee of the Southwest Hospital (ChiCTR-DCC-11001733).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 2013;132:1133-45. [Crossref] [PubMed]
- Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett 1996;392:49-53. [Crossref] [PubMed]
- Wang Z, Zhang X, Shen P, et al. Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochem Biophys Res Commun 2005;336:1023-7. [Crossref] [PubMed]
- Shi L, Dong B, Li Z, et al. Expression of ER-{alpha}36, a novel variant of estrogen receptor {alpha}, and resistance to tamoxifen treatment in breast cancer. J Clin Oncol 2009;27:3423-9. [Crossref] [PubMed]
- Lewis JS, Jordan VC. Selective estrogen receptor modulators (SERMs): mechanisms of anticarcinogenesis and drug resistance. Mutat Res 2005;591:247-63. [Crossref] [PubMed]
- Lin SL, Yan LY, Zhang XT, et al. ER-alpha36, a variant of ER-alpha, promotes tamoxifen agonist action in endometrial cancer cells via the MAPK/ERK and PI3K/Akt pathways. PLoS One 2010;5:e9013 [Crossref] [PubMed]
- Wang Q, Jiang J, Ying G, et al. Tamoxifen enhances stemness and promotes metastasis of ERα36(+) breast cancer by upregulating ALDH1A1 in cancer cells. Cell Res 2018;28:336-58. [Crossref] [PubMed]
- Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen receptor beta: an overview and update. Nucl Recept Signal 2008;6:e003 [PubMed]
- Ogawa S, Inoue S, Watanabe T, et al. The complete primary structure of human estrogen receptor beta (hER beta) and its heterodimerization with ER alpha in vivo and in vitro. Biochem Biophys Res Commun 1998;243:122-6. [Crossref] [PubMed]
- Leung YK, Mak P, Hassan S, et al. Estrogen receptor (ER)-beta isoforms: a key to understanding ER-beta signaling. Proc Natl Acad Sci U S A 2006;103:13162-7. Erratum in: Proc Natl Acad Sci U S A 2006;103:14977. [Crossref] [PubMed]
- Chen L, Qiu J, Yang C, et al. Identification of a novel estrogen receptor beta1 binding partner, inhibitor of differentiation-1, and role of ERbeta1 in human breast cancer cells. Cancer Lett 2009;278:210-9. [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Chang HG, Kim SJ, Chung KW, et al. Tamoxifen-resistant breast cancers show less frequent methylation of the estrogen receptor beta but not the estrogen receptor alpha gene. J Mol Med (Berl) 2005;83:132-9. [Crossref] [PubMed]
- Guo L, Meng J, Yilamu D, et al. Significance of ERβ expression in different molecular subtypes of breast cancer. Diagn Pathol 2014;9:20. [Crossref] [PubMed]
- Omoto Y, Inoue S, Ogawa S, et al. Clinical value of the wild-type estrogen receptor beta expression in breast cancer. Cancer Lett 2001;163:207-12. [Crossref] [PubMed]
- Järvinen TA, Pelto-Huikko M, Holli K, et al. Estrogen receptor beta is coexpressed with ERalpha and PR and associated with nodal status, grade, and proliferation rate in breast cancer. Am J Pathol 2000;156:29-35. [Crossref] [PubMed]
- Madeira M, Mattar A, Logullo AF, et al. Estrogen receptor alpha/beta ratio and estrogen receptor beta as predictors of endocrine therapy responsiveness-a randomized neoadjuvant trial comparison between anastrozole and tamoxifen for the treatment of postmenopausal breast cancer. BMC Cancer 2013;13:425. [Crossref] [PubMed]
- Marotti JD, Collins LC, Hu R, et al. Estrogen receptor-beta expression in invasive breast cancer in relation to molecular phenotype: results from the Nurses' Health Study. Mod Pathol 2010;23:197-204. [Crossref] [PubMed]
- Thomas C, Gustafsson JÅ. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer 2011;11:597-608. [Crossref] [PubMed]
- Thomas C, Rajapaksa G, Nikolos F, et al. ERbeta1 represses basal breast cancer epithelial to mesenchymal transition by destabilizing EGFR. Breast Cancer Res 2012;14:R148. [Crossref] [PubMed]
- Shanle EK, Zhao Z, Hawse J, et al. Research resource: global identification of estrogen receptor β target genes in triple negative breast cancer cells. Mol Endocrinol 2013;27:1762-75. [Crossref] [PubMed]
- Cotrim CZ, Fabris V, Doria ML, et al. Estrogen receptor beta growth-inhibitory effects are repressed through activation of MAPK and PI3K signalling in mammary epithelial and breast cancer cells. Oncogene 2013;32:2390-402. [Crossref] [PubMed]
- Honma N, Horii R, Iwase T, et al. Clinical importance of estrogen receptor-beta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J Clin Oncol 2008;26:3727-34. [Crossref] [PubMed]
- Clark GM, Osborne CK, McGuire WL. Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J Clin Oncol 1984;2:1102-9. [Crossref] [PubMed]
- Wang Z, Zhang X, Shen P, et al. A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc Natl Acad Sci U S A 2006;103:9063-8. [Crossref] [PubMed]
- Kang L, Zhang X, Xie Y, et al. Involvement of estrogen receptor variant ER-alpha36, not GPR30, in nongenomic estrogen signaling. Mol Endocrinol 2010;24:709-21. [Crossref] [PubMed]
- Lindberg K, Helguero LA, Omoto Y, et al. Estrogen receptor β represses Akt signaling in breast cancer cells via downregulation of HER2/HER3 and upregulation of PTEN: implications for tamoxifen sensitivity. Breast Cancer Res 2011;13:R43. [Crossref] [PubMed]
- Wang J, Zhang C, Chen K, et al. ERβ1 inversely correlates with PTEN/PI3K/AKT pathway and predicts a favorable prognosis in triple-negative breast cancer. Breast Cancer Res Treat 2015;152:255-69. [Crossref] [PubMed]


