
Genetic signatures on prostate biopsy: clinical implications
Introduction
Prostate cancer is the most common non-cutaneous malignancy in men with 161,000 new diagnoses in 2017 (1). It is a very heterogeneous disease with variable outcomes, depending on disease stage and grade. Due to serum prostate-specific antigen (PSA) screening of men at risk, most patients are diagnosed with indolent disease that does not impact quality of life or life expectancy and is therefore managed expectantly [active surveillance (AS)]. However, some patients will die of their cancer if left untreated.
Different clinical and pathologic factors, such as PSA level, tumor stage and grade, and presence or absence of lymph node metastasis significantly correlate with the prognosis of the underlying disease. One of the most significant prognostic parameters is the Gleason biopsy grading system (2), comprising five histologic grades of prostate adenocarcinoma: Gleason pattern 1 to 5. Pattern 1 represents the best differentiated, i.e., it is the closest in histologic appearance to benign prostate gland, while pattern 5 is the least differentiated and most aggressive type of prostate adenocarcinoma. The sum of the two most prevalent Gleason patterns equals the Gleason score (GS). Nowadays, a GS of ≤5 (primary + secondary Gleason pattern) is rarely described, as Gleason patterns 1 and 2 are histomorphologically almost indistinguishable from normal prostate tissue. The most recent modification to this pathological schema is the introduction of grade groups based on the GS, reorienting the numerical system to more accurately reflect the aggressiveness on a scale from 1 to 5 (3). The grade group allows for better discrimination at the lower grade of prostate pathology, specifically for GS 7 which can now be separated into grade group 2 (3+4) or grade group 3 (4+3). This simple modification holds the potential to help improve practices regarding prostate cancer management by highlighting the subtle differences in pathology that might have an impact on prognosis (4).
Microscopic assessment and quantification of Gleason grade is somewhat subjective, and there is evidence that random prostate biopsies might under-sample, and hence under-stage and -grade the existing cancer. Therefore, additional genetic and genomic tools to better characterize the cancer biology are being studied. Genetic testing in prostate cancer (i.e., the study of individual up- or down-regulated genes that have been found to be associated with the grade of the tumor) is important as family predisposition is responsible for about 5–15% of all prostate cancers (5-7). In contrast, genomic studies assess all genes in the genome and their interactions that can directly influence the biology and behavior of the tumor itself; studying these activities can then help to more objectively and independently assess underlying risk of undetected prostate cancer, as well as the tumor grade in patients found to have cancer on prostate biopsy.
The reason why this is so important is because most very low and low risk prostate cancers are, depending on the individual circumstances, age, and life expectancy of the patient, not necessarily being treated nowadays, but closely monitored (i.e., AS). In contrast, localized but more aggressive (intermediate or high-risk) cancers are usually being referred to definitive curative treatment options. Distinguishing the different types of cancers through histological, genetic and genomic analyses therefore helps counseling and reassuring patients, and guiding patients toward the appropriate and necessary management route.
This review is aimed at giving an overview of the different types of genetic and genomic tests that exist to determine the risk of the presence of non-indolent prostate cancer on prostate biopsy, i.e., to determine the necessity for the performance of a prostate biopsy or to repeat a biopsy, and to examine prostate biopsy tissue for further characterization of the cancer biology to aid in the decision-making process with regards to surveillance or treatment in clinical practice.
Prostate cancer genetics
The genetic drivers underlying prostate carcinoma oncogenesis provide information useful for diagnostic, prognostic, and therapeutic purposes. The early efforts within this advanced diagnostic area used immunhistochemical (IHC) staining to determine the level of expression of a specific gene within a pathological specimen. One such study found that expression of p27, a cell cycle regulator, negatively correlated with biochemical recurrence (BCR), with tumors staining at <45% showing a 2.5-fold increase in the risk of BCR (8). Although IHC is still widely used for prostate cancer research and diagnosis [i.e., AMACR, androgen receptor (AR), PSA, etc.], the widespread availability and dwindling costs associated with genetic sequencing techniques offers a more precise assessment of a tumor’s mutational profile.
The landmark publication of the Cancer Genome Atlas for prostate carcinoma in 2015 used whole-exome sequencing to develop a molecular taxonomy of 7 distinct subgroups of genetic mutations into which 74% of all tumors examined would be classified (9). For example, overexpression of the E26 transformation-specific (ETS) family of oncogenes, regulated by androgen-regulated stimulation, is present in over 50% of tumors. This overexpression was found to be due to chromosomal rearrangement resulting in fusion of the promoter TMPRSS2 to an ETS gene, most often ERG (10). There are no commercially available therapeutic agents available to date that have been able to take advantage of TMPRSS2-ERG rearrangement, however, its high overall prevalence has led to the development of several prognostic tests based on its detection, as will be discussed in subsequent sections.
There are more recent efforts, beyond simply identifying genetic mutations present in prostate cancer, which seek to evaluate clonal evolution within the lifespan of a tumor (11-13). This is particularly important when considering ideal tests for early detection and prognostication, since a mutation present in metastatic disease may not be present in the beginning stages of localized disease. In these circumstances, when such a mutation is detected in a patient otherwise lacking clinical signs or symptoms of distant spread, it may indicate a need for more aggressive treatment, independent of other factors (i.e., PSA, GS, etc.).
Pre-prostate biopsy decision making
The availability of genetic testing comes at the earliest stages of prostate cancer diagnosis when the decision of whether or not to proceed with a prostate biopsy is being contemplated. Table 1 shows an overview of non-tissue based genomic tests. Given the intense scrutiny surrounding over-diagnosis in prostate cancer, among other malignancies, there is a demand for increased sensitivity and specificity in our screening tests. Like all commercially available genetic testing for prostate cancer, these tests are prognostic, offering insight into which patients might benefit most from a more aggressive therapeutic strategy rather than identifying actionable mutations for targeted treatment. In reviewing the available literature on this topic, it becomes clear that the development of each test has followed a very similar pathway: (I) identifying a cohort of patients with clinically significant prostate cancer or certain adverse clinicopathologic features; (II) genetic sequencing of prostatectomy specimens, prostate biopsies, and/or bodily fluids in these patients to identify mutations, or combinations of mutations, that appear more frequently than in matched benign controls; (III) experimental validation of the newly identified biomarkers to ensure feasibility, and, finally; (IV) clinical validation in a cohort with sufficient follow-up to draw significant conclusions regarding the prognostic ability of the test.
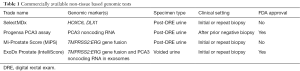
Full table
SelectMDx
The SelectMDx test was developed by MDxHealth (Irvine, CA, USA) as a urinary biomarker collected in the first voided urine specimens after digital rectal exam of the prostate (DRE). In the exploratory analysis, the biomarker discovery phase identified 39 potential prostate cancer biomarkers using gene expression profiling data from the transurethral resection of prostate (TURP) or radical prostatectomy (RP) specimens of 133 men (14). The pathology within this cohort included normal prostate, benign prostatic hyperplasia (BPH), low-grade prostate cancer (GS ≤6), high-grade prostate cancer (GS ≥7), castration-resistant prostate cancer (CRPC), and metastatic prostate cancer. Using a separate cohort of men undergoing initial or repeat prostate biopsy for elevated PSA, the researchers identified a panel of 3 genes (HOXC6, TDRD1, and DLX1) that served to increase the AUC for predicting presence of GS ≥7 when compared to PCA3 or PSA alone (AUC 0.77 vs. 0.68 vs. 0.72, respectively). When clinically validated, the HOXC6 and DLX1 expression profiles had the best combination of performance (AUC 0.73) and analytic reproducibility (15). In combination with other clinicopathologic features (PSA, PSA density, age, family history, and any prior prostate biopsy results) the AUC improved to 0.90. It is worth noting that this was only slightly better than the predictive ability of the clinicopathologic features alone which had an AUC of 0.87, though the difference was found to be statistically significant (P=0.018). To date, SelectMDx is still under consideration for FDA approval.
PCA3
Prostate cancer antigen 3 (PCA3) is a noncoding RNA from chromosome 9p21-22 that was found to be overexpressed by 10- to 100-fold in the tissue of men with prostate cancer (16). The Progensa PCA3 score available from Hologic, Inc. (Marlborough, MA, USA) calculated by measuring the level of PCA3 mRNA as a ratio to PSA mRNA within first voided urine sample after brief prostate massage, and has been shown to improve upon the predictive ability of serum PSA for finding any prostate cancer at the time of prostate biopsy (17-22). The critical validation study was conducted in 1,140 men within the placebo arm of the REDUCE trial who provided post-DRE urine samples prior to their 2- and 4-year per protocol prostate biopsies. Given that all men were required to have a negative prostate biopsy at study entry this represented an entirely repeat biopsy cohort (17). While the median PCA3 score was higher for GS ≥7 (49.5) compared to GS 6 (31.8), there was no statistically significant difference between the AUC for high- and low-grade disease. PCA3 score performed the best when used as part of a model including clinical factors (PSA, percent free PSA, prostate volume, age, and family history) with an AUC of 0.753 compared to 0.612 for PSA alone when the cutoff value was set to >35 when the PSA was between 2.5 and 10 ng/dL. FDA approval is limited to the repeat biopsy setting in large part due to the results of subsequent studies that have shown unacceptably high rates of missed GS ≥7 cancers when applied to biopsy-naïve patients; upwards of 13% on initial biopsy (18,19). Recently, Wei et al. (22) argued that PCA3 can still be useful for ruling-in an initial biopsy with a higher cutoff value of >60 when the PSA results are otherwise equivocal.
MiPS
By combining the most commonly identified genetic mutation found in prostate cancer, the TMPRSS2:ERG gene fusion (10), with PCA3, the University of Michigan MLab (Ann Arbor, MI, USA) introduced the Mi-Prostate Score (MiPS) which measures the expression of each within a urine sample. Unlike PCA3 alone, MiPS has shown utility both in the initial and repeat biopsy setting by outperforming PSA alone, as well as the ERSPC and PCPT risk calculators (23,24). Sensitivity was measured at 91% for detecting GS ≥7 in a cohort that was comprised largely of biopsy-naïve patients (79%), with an AUC 0.842 if combined with the ERSPC risk calculator (23). In an AS cohort, increasing MiPS, measured as a continuous variable, was significantly associated with negative biopsy outcome, GS 6, and GS ≥7 cancer (25). Though it is not yet FDA-approved, this test holds potential for reducing the number of unnecessary biopsies by approximately 50% while maintaining good discrimination for high-grade disease (24,26).
ExoDx Prostate (IntelliScore)
Urinary exosomes are small, bilipid walled vesicles that are shed by cells and can be found in various bodily fluids, including prostatic secretions, while containing highly enriched levels of mRNA compared to total cell RNA (27,28). The Prostate (IntelliScore) from Exome Diagnostics (Waltham, MA, USA) is an FDA-approved test which capitalizes on the fact that both TMPRSS2:ERG and PCA3 are found at levels nearly 100-fold higher within urinary exosomes than post-DRE urine specimens (28). Clinical validation comes by way of a 2016 study (29) comprised of a training cohort (n=499) and validation cohort (n=1,064) with the specific aim of predicting detection of GS ≥7 with the specific aim of predicting detection of GSn cohort ng/dL. The AUC for exosomes plus clinical parameters (PSA, age, race, and family history) was 0.73 compared to 0.55 for PSA alone. The test showed high sensitivity and negative predictive value (NPV) (92% and 91%, respectively) and led to a 27% reduction in the number of prostate biopsies, however, the tradeoff was that 8% of GS ≥7 cancers would be missed using this method.
It is worth noting that while all of the aforementioned genetic biomarker tests have demonstrated statistical improvement in prostate cancer detection and/or discrimination of higher grade disease, there have been no published studies to date on the cost-effectiveness of these approaches. In real-world scenarios, insurers will demand to see not only clinical efficacy but also a concomitant change in practice patterns that result in significant cost-savings.
Prostate biopsy based genetic testing
There are definite advantages to non-invasive genetic testing with regards to patient comfort and inconvenience, however, tissue samples are still required for more detailed genomic analysis at present time (Table 2). The prostate biopsy based tests require a small section of diagnostic material to be sent to a central laboratory for analysis, relying upon the services of local pathologists to select an appropriate sample of tissue. The raw data generated from genetic testing is then interpreted in the context of the validation cohorts for each specific product and then reported to the physician and patient in an easy to read manner that typically provides some type of “score” with a corresponding percentage of patients who developed a specific outcome (i.e., BCR, metastasis, overall survival).

Full table
ConfirmMDx
The ConfirmMDx test was also developed by MDxHealth (Irvine, CA, USA). It is a diagnostic tissue-based test to detect occult prostate cancer in histopathologically negative prostate biopsy tissue, and is mainly based on the “field-effect” (i.e., changes in tissues surrounding cancerous lesions) of epigenetic hypermethylation changes of three genes (GSTP1, APC, RASSF).
Originally described by Lee et al. in 1994, epigenetic hypermethylation mutations of regulatory sequences of the pi-class glutathione S-transferase gene (GSTP1) were found to be highly prevalent only in prostate cancer, but not in benign tissue (30). A subsequent cohort study assessing epigenetic changes in prostate cancer tissue confirmed these findings, and was used for the development and optimization of an epigenetic multiplex assay based on the above three genes (31). Two subsequent retrospective clinical validation studies confirmed the predictive accuracy of hypermethylation changes on prostate cancer-negative biopsy tissue for the presence of significant prostate cancer on repeat biopsy: In the MATLOC (Methylation Analysis to Locate Occult Cancer) study, 483 men in the UK underwent ConfirmMDx testing on initial biopsy; this analysis found a sensitivity of 68%, a specificity of 64%, and a NPV of 90% for the absence of cancer on repeat prostate biopsy samples. Interestingly, all cases with a GS of ≥8 tested positive for epigenetic hypermethylation (32). The DOCUMENT (Detection Of Cancer Using Methylated Events in Negative Tissue) trial, a validation study of the MATLOC analysis in a US cohort of men, confirmed these findings, and hypermethylation mutations of regulatory sequences of GSTP1 were again found to be the most accurate predictor for the presence of prostate cancer on repeat biopsy after initial negative biopsy (33).
Detecting areas that test positive for hypermethylation in the identified genes aids in the identification of biopsies with false-negative histopathological results, and might help in the decision-making whether a repeat biopsy is warranted. Consequently, ConfirmMDx test might help decrease the number of unnecessary repeat prostate biopsies. It was incorporated into the NCCN guidelines in 2016 (34).
OncotypeDx Genomic Prostate Score (GPS)
Since 2004, Genomic Health, Inc. (Redwood City, CA, USA) has developed a variety of validated multi-gene real-time polymerase chain reaction (RT-PCR) assays to evaluate the individual underlying cancer biology in patients with different types of cancers, i.e., breast, colon, prostate. The Oncotype DX Prostate Cancer Assay platform is able to examine small amounts of prostate tissue for the expression patterns of 12 cancer-related genes, representing 4 distinct biological pathways in prostate tumorigenesis (stromal response: BGN, COL1A1, SFRP4; cellular organization: FLNC, GSN, TPM2, GSTM2; androgen pathway: FAM13C, KLK2, AZGP1, SRD5A2; proliferation: TPX2). It also includes 5 reference genes (ARF1, ATP5E, CLTC, GPS1 and PGK1). Together, this 17-gene assay has been shown in analytical validation studies to be able to reproducibly calculate the GPS, a score that in numerous subsequent clinical validation studies was confirmed to correlate with adverse pathologic findings on RP specimens and was also able to predict risk of BCR (35-37). Even in non-cancerous regions in prostatectomy specimens of patients that underwent removal of the prostate for biopsy-proven cancer, the GPS showed similar performance characteristics as in cancerous lesions themselves, suggesting that a “field effect” within the prostate is present as well, as was described earlier (38). A prospective observational study of community-based urology practices in the US examining the use of GPS in the decision-making process of patients with newly-diagnosed prostate cancer is currently being conducted. Preliminary data from this study on the first 258 enrolled patients after one year of follow-up found that men were more likely to choose AS if GPS was used, in comparison to a control cohort in which GPS was not used (62% vs. 40%). Overall, 55% of patients who underwent GPS elected AS and were still on AS at one year, compared to 34% of the control group. The rate of patients that continued on AS at the one-year time point was similar between the two groups (89% vs. 86%) (39).
In summary, Oncotype DX GPS helps assess underlying tumor biology on prostate biopsy tissue, and has a significant impact in the decision-making between initial treatment or surveillance options for patients with newly-diagnosed prostate cancer. Whether it also has a significant benefit during the course of surveillance remains to be established.
Prolaris
Cancer cells exhibit higher levels of expression from certain cell cycle related genes when compared to their benign counterparts leading to unregulated cellular proliferation, a hallmark of cancer oncogenesis (40). Expression levels from 874 candidate genes were quantified in HeLa cells at various time points as they progressed through the cell cycle in some of the early pioneering work in this field of study. When compared to benign cells, the HeLa cells showed higher levels of those involved in DNA replication and chromosomal segregation. The Prolaris test from Myriad Genetics (Salt Lake City, UT, USA) has taken advantage of these observations to develop a 46-gene cell cycle progression (CCP) score for predicting adverse outcomes from prostate biopsy samples (41).
This test was originally developed and validated for use on RP specimens as a way to improve risk stratification post-operatively and predict BCR (42,43). Of those 874 candidate genes identified by Whitfield et al. (40), 31 were selected for use in the CCP score due to their increased level of expression in patients with the adverse outcomes of interest, while an additional 15 genes that exhibited constant expression were used as internal controls (43). In this post-RP setting the 10-year risk of mortality based on AUA risk category alone was 4.8%, but adding the CCP score widened the discriminatory ability of the test from 1.8% to 6.7%. The authors then took this concept a step further and used the CCP score on prostate biopsy specimens where it was expected to make a greater impact in the decision-making process, before a definitive therapy had been carried out. Using core-needle biopsies from 349 “conservatively treated” men with prostate cancer from a UK registry, the CCP score was able to significantly predict prostate cancer mortality with a HR 1.65 per one unit increase (44). Furthermore, the 10-year cancer specific mortality risk could be reported as a percentage based on CCP score, ranging from 19% for score of <0 up to 74.9% for scores of 3 or greater. Subsequent studies have further validated these findings with HR for disease specific mortality on multivariate analysis (controlling for GS, PSA, clinical stage, etc.) ranging from 1.47 to 2.17 per unit increase (41,42). Prolaris offers the potential of identifying men at both increased and decreased risk of prostate cancer death, as shown by Cuzick and colleagues where 14% of the CAPRA low risk (<4% 10-year cancer specific mortality) patients were reclassified into higher-risk groups and 44% of CAPRA intermediate-risk patients were downgraded to low-risk, based on the results of the CCP score (41).
The ability of the Prolaris test to change clinical practice has been published more than any other prostate biopsy based genomic test currently available (45-47). In one such study, physicians were surveyed regarding their pre- and post-test treatment decisions for 331 patients diagnosed with prostate cancer on needle biopsy (45). The Polaris test influenced physicians in at least some way for 97.8% of patients and led to an overall reduction in the therapeutic burden, as measured by change from interventional to non-interventional treatment strategies for this predominantly low-grade (80.1% grade group 1 or 2) cohort. Though the authors found 80.2% concordance with the ultimate treatment delivered, the conclusions drawn from a survey-based study should be considered with a great deal of skepticism. In a larger cohort, Shore et al. (47) found that the there was no significant difference in the recommendation for nonintervention between pre- and post-Prolaris test result treatment plans. Rather, what was observed was a “reshuffling” of patients from the intervention and non-intervention groups based on the Prolaris test, creating no net difference. The rate of RP declined by 34% from pre- to post-test, however, attributing such a change to the test alone is impossible given the numerous other potential factors that could be implicated in the ultimate treatment decision. While such studies may be useful in garnering opinions of physicians using these tests under “real-world” conditions, more objective endpoints (cost-benefit, survival, etc.) are required to truly evaluate clinical impact.
Decipher
The Decipher test (GenomeDx, Vancouver, BC, CA) was originally conceived as a prostatectomy based tissue assay for predicting early metastasis in men with high-risk features (pT3, positive margins, GS ≥8, PSA >20 ng/mL) (48). The initial discovery and validation cohort specifically included men with regional and/or distant metastasis within 5 years of BCR for comparison against those with no evidence of disease recurrence (PSA or other signs) for at least 7 years post-operatively (49). Whole transcriptome RNA microarray and statistical regressions yielded 22 genomic markers from both coding and non-coding regions of the genome to be included in the so-called “genomic classifier” (GC).
Clinical validation in RP specimens showed that the Decipher score outperforms CAPRA-S for the prediction of early metastatic disease, though the absolute difference is small (c-index 0.75 vs. 0.72) (50). Using the same cohort of patients, Klein et al. (48) applied Decipher to the pre-RP biopsies and compared it to the NCCN risk stratification model (34). The Decipher score alone had a c-index of 0.80 for predicting 10-year post-RP risk of metastasis compared to 0.75 for NCCN risk group category alone; this increased to 0.88 when both Decipher and NCCN were combined. In another biopsy study, each 10% increase in the Decipher score correlated with an increase in risk of metastasis with a HR of 1.53 (51). Prostate cancer specific mortality was not a primary endpoint of this study. However, low- and intermediate-risk Decipher scores correlated with a 0% chance of prostate cancer death within 5-year of receiving definitive therapy.
Prostate Core Mitomic Test
The Prostate Core Mitomic Test (PCMT) from MDNA Life Sciences (West Palm Beach, FL, USA) is based on the “tumor field effect” of prostate cancer and specifically focuses on alterations of the mitochondrial genome (52). Preliminary studies showed that benign tissue obtained from RP specimens containing foci of prostate cancer exhibited equal mutation rate of the mitochondrial genome from all areas sampled, regardless of the pathologic diagnosis (53). This test is commercially available but not FDA-approved as of yet, with minimal published data available. The clinical validation study used men with negative prostate biopsies who underwent repeat biopsy within 1 year, finding a NPV of 91% with AUC 0.75 on the second specimen (54).
Limitations
Definite statistical improvements in prostate cancer prognosis have been reported widely throughout the literature, however, these tests are far from ideal and carry with them a few important limitations to keep in mind when applying them in clinical practice. The biopsy procedure itself is, in fact, a limitation to the application of prostate cancer genomics, much in the same way it limits traditional pathologic diagnosis. When biopsy specimens are compared to the final RP specimen, the tumor grade will be discordant for 25% to 50% of patients, mainly due to biopsy sampling error (55-57). This means that the pathologist will often not be presented with a sample from the most relevant region (i.e., highest GS) based on 12 tissue cores alone. Genomic testing may provide an advantage in this regard since genetic alterations likely precede phenotypic changes detectable on microscopy, but the degree to which all tumor foci are genetically linked is still debatable (polyclonality vs. field cancerization) (36,52,58,59). Thus, questions about the role of sampling error persists in prostate-biopsy based genomic tests.
Interobserver variability in Gleason scoring has been well documented but genetic testing offers the theoretical promise of true objectivity because it no longer depends on human interpretation (56,60). However, the tissue-based tests require representative sections to be selected by local pathologists and sent to a central location for expression profiling, introducing some degree of error between individuals. Furthermore, the genetic tests themselves reportedly yielded non-diagnostic results in approximately 10–20% of samples for each of the validation studies previously mentioned (15,17,29,36,48,61).
Finally, perhaps the most significant factor limiting clinical utility of prostate cancer genomics is the lack of available cost-effectiveness data. There have been several studies examining self-reported changes in physician practice habits (39,45-47,62), but no large-scale analysis of the real-world impact that these tests are having on patient outcomes. There is a hypothesized benefit from gaining additional prognostic information but, on the other hand, without evidence of such benefits (i.e., improved survival, reduced overall cost of treatment, etc.) it is unclear how the currently available products are going to fit into contemporary and future prostate cancer management.
Future directions
Genetic sequencing is no longer restricted to highly specialized laboratories at major universities and the widespread availability of inexpensive next-generation sequencing platforms is sure to generate additional valuable data that will impact the way we care for prostate cancer. The literature is full of publications containing potential future biomarkers touting improvement in risk stratification and higher diagnostic accuracy (63-65). Ultimately, the ideal prostate biopsy based test would not only offer prognostic insights but also provide therapeutic targets to improve disease outcomes. In much the same way as the AR-V7 splice variant identifies men who are unlikely to benefit from enzalutamide therapy (66) or SPOP mutation predicts response to PARP-inhibitors (67), genomic information from biopsy specimens could determine suitability for radiotherapy versus RP (68-70).
Conclusions
Prostate cancer management has only begun to integrate genomic biomarkers despite their use for over a decade in other cancers, specifically breast. The tests available at the present time reflect the unmet need for better discrimination for adverse disease outcomes in clinical risk-stratification models and aim to reduce the burden of overtreatment while identifying patients who would benefit most from early, aggressive intervention. The current landscape represents an incremental improvement over the previously available tools based on analysis of retrospective cohorts, but prospectively generated outcomes data will take many more years to mature.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Israel Deutsch, James McKiernan, Charles Drake) for the series “Prostate Cancer: Current Understanding and Future Directions” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.21). The series “Prostate Cancer: Current Understanding and Future Directions” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep 1966;50:125-8. [PubMed]
- Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 2016;40:244-52. [PubMed]
- Thomsen FB, Folkvaljon Y, Brasso K, et al. Prognostic implications of 2005 Gleason grade modification. Population-based study of biochemical recurrence following radical prostatectomy. J Surg Oncol 2016;114:664-70. [Crossref] [PubMed]
- Benafif S, Eeles R. Genetic predisposition to prostate cancer. Br Med Bull 2016;120:75-89. [Crossref] [PubMed]
- Carter BS, Beaty TH, Steinberg GD, et al. Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci U S A 1992;89:3367-71. [Crossref] [PubMed]
- Zeegers MP, Jellema A, Ostrer H. Empiric risk of prostate carcinoma for relatives of patients with prostate carcinoma: a meta-analysis. Cancer 2003;97:1894-903. [Crossref] [PubMed]
- Freedland SJ, deGregorio F, Sacoolidge JC, et al. Preoperative p27 status is an independent predictor of prostate specific antigen failure following radical prostatectomy. J Urol 2003;169:1325-30. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011-25. [Crossref] [PubMed]
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644-8. [Crossref] [PubMed]
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77. [Crossref] [PubMed]
- Gumuskaya B, Gurel B, Fedor H, et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis 2013;16:209-15. [Crossref] [PubMed]
- Rodrigues DN, Boysen G, Sumanasuriya S, et al. The molecular underpinnings of prostate cancer: impacts on management and pathology practice. J Pathol 2017;241:173-82. [Crossref] [PubMed]
- Leyten GH, Hessels D, Smit FP, et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res 2015;21:3061-70. [Crossref] [PubMed]
- Van Neste L, Hendriks RJ, Dijkstra S, et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur Urol 2016;70:740-8. [Crossref] [PubMed]
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59:5975-9. [PubMed]
- Aubin SM, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol 2010;184:1947-52. [Crossref] [PubMed]
- Chevli KK, Duff M, Walter P, et al. Urinary PCA3 as a predictor of prostate cancer in a cohort of 3,073 men undergoing initial prostate biopsy. J Urol 2014;191:1743-8. [Crossref] [PubMed]
- de la Taille A, Irani J, Graefen M, et al. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J Urol 2011;185:2119-25. [Crossref] [PubMed]
- Hessels D, van Gils MP, van Hooij O, et al. Predictive value of PCA3 in urinary sediments in determining clinico-pathological characteristics of prostate cancer. Prostate 2010;70:10-6. [Crossref] [PubMed]
- Marks LS, Fradet Y, Deras IL, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 2007;69:532-5. [Crossref] [PubMed]
- Wei JT, Feng Z, Partin AW, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol 2014;32:4066-72. [Crossref] [PubMed]
- Leyten GH, Hessels D, Jannink SA, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol 2014;65:534-42. [Crossref] [PubMed]
- Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur Urol 2016;70:45-53. [Crossref] [PubMed]
- Lin DW, Newcomb LF, Brown EC, et al. Urinary TMPRSS2:ERG and PCA3 in an active surveillance cohort: results from a baseline analysis in the Canary Prostate Active Surveillance Study. Clin Cancer Res 2013;19:2442-50. [Crossref] [PubMed]
- Carlsson SV, Roobol MJ. Improving the evaluation and diagnosis of clinically significant prostate cancer in 2017. Curr Opin Urol 2017;27:198-204. [Crossref] [PubMed]
- Mitchell PJ, Welton J, Staffurth J, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med 2009;7:4. [Crossref] [PubMed]
- Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100:1603-7. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, O'Neill V, et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol 2016;2:882-9. [Crossref] [PubMed]
- Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A 1994;91:11733-7. [Crossref] [PubMed]
- Van Neste L, Bigley J, Toll A, et al. A tissue biopsy-based epigenetic multiplex PCR assay for prostate cancer detection. BMC Urol 2012;12:16. [Crossref] [PubMed]
- Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol 2013;189:1110-6. [Crossref] [PubMed]
- Partin AW, Van Neste L, Klein EA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192:1081-7. [Crossref] [PubMed]
- Carroll PR, Parsons JK, Andriole G, et al. NCCN Guidelines Insights: Prostate Cancer Early Detection, Version 2.2016. J Natl Compr Canc Netw 2016;14:509-19. [Crossref] [PubMed]
- Cullen J, Rosner IL, Brand TC, et al. A Biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol 2015;68:123-31. [Crossref] [PubMed]
- Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol 2014;66:550-60. [Crossref] [PubMed]
- Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics 2013;14:690. [Crossref] [PubMed]
- Magi-Galluzzi C, Maddala T, Falzarano SM, et al. Gene expression in normal-appearing tissue adjacent to prostate cancers are predictive of clinical outcome: evidence for a biologically meaningful field effect. Oncotarget 2016;7:33855-65. [Crossref] [PubMed]
- Eure G, Germany R, Given R, et al. Use of a 17-Gene Prognostic Assay in Contemporary Urologic Practice: Results of an Interim Analysis in an Observational Cohort. Urology 2017;107:67-75. [Crossref] [PubMed]
- Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 2002;13:1977-2000. [Crossref] [PubMed]
- Cuzick J, Stone S, Fisher G, et al. Validation of an RNA cell cycle progression score for predicting death from prostate cancer in a conservatively managed needle biopsy cohort. Br J Cancer 2015;113:382-9. [Crossref] [PubMed]
- Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol 2013;31:1428-34. [Crossref] [PubMed]
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245-55. [Crossref] [PubMed]
- Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer 2012;106:1095-9. [Crossref] [PubMed]
- Crawford ED, Scholz MC, Kar AJ, et al. Cell cycle progression score and treatment decisions in prostate cancer: results from an ongoing registry. Curr Med Res Opin 2014;30:1025-31. [Crossref] [PubMed]
- Shore N, Concepcion R, Saltzstein D, et al. Clinical utility of a biopsy-based cell cycle gene expression assay in localized prostate cancer. Curr Med Res Opin 2014;30:547-53. [Crossref] [PubMed]
- Shore ND, Kella N, Moran B, et al. Impact of the cell cycle progression test on physician and patient treatment selection for localized prostate cancer. J Urol 2016;195:612-8. [Crossref] [PubMed]
- Klein EA, Haddad Z, Yousefi K, et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology 2016;90:148-52. [Crossref] [PubMed]
- Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 2013;8:e66855 [Crossref] [PubMed]
- Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol 2015;67:778-86. [Crossref] [PubMed]
- Nguyen PL, Haddad Z, Ross AE, et al. Ability of a Genomic classifier to predict metastasis and prostate cancer-specific mortality after radiation or surgery based on needle biopsy specimens. Eur Urol 2017;72:845-52. [Crossref] [PubMed]
- Maki J, Robinson K, Reguly B, et al. Mitochondrial genome deletion aids in the identification of false- and true-negative prostate needle core biopsy specimens. Am J Clin Pathol 2008;129:57-66. [Crossref] [PubMed]
- Parr RL, Dakubo GD, Crandall KA, et al. Somatic mitochondrial DNA mutations in prostate cancer and normal appearing adjacent glands in comparison to age-matched prostate samples without malignant histology. J Mol Diagn 2006;8:312-9. [Crossref] [PubMed]
- Robinson K, Creed J, Reguly B, et al. Accurate prediction of repeat prostate biopsy outcomes by a mitochondrial DNA deletion assay. Prostate Cancer Prostatic Dis 2010;13:126-31. [Crossref] [PubMed]
- Davies JD, Aghazadeh MA, Phillips S, et al. Prostate size as a predictor of Gleason score upgrading in patients with low risk prostate cancer. J Urol 2011;186:2221-7. [Crossref] [PubMed]
- Epstein JI, Feng Z, Trock BJ, et al. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol 2012;61:1019-24. [Crossref] [PubMed]
- Kvåle R, Moller B, Wahlqvist R, et al. Concordance between Gleason scores of needle biopsies and radical prostatectomy specimens: a population-based study. BJU Int 2009;103:1647-54. [Crossref] [PubMed]
- Cooper CS, Eeles R, Wedge DC, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet 2015;47:367-72. [Crossref] [PubMed]
- Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736-45. [Crossref] [PubMed]
- Goodman M, Ward KC, Osunkoya AO, et al. Frequency and determinants of disagreement and error in gleason scores: a population-based study of prostate cancer. Prostate 2012;72:1389-98. [Crossref] [PubMed]
- Bishoff JT, Freedland SJ, Gerber L, et al. Prognostic utility of the cell cycle progression score generated from biopsy in men treated with prostatectomy. J Urol 2014;192:409-14. [Crossref] [PubMed]
- Badani KK, Kemeter MJ, Febbo PG, et al. The impact of a biopsy based 17-gene genomic prostate score on treatment recommendations in men with newly diagnosed clinically prostate cancer who are candidates for active surveillance. Urol Pract 2015;2:181-9. [Crossref]
- Albitar M, Ma W, Lund L, et al. Predicting Prostate biopsy results using a panel of plasma and urine biomarkers combined in a Scoring System. J Cancer 2016;7:297-303. [Crossref] [PubMed]
- Lalonde E, Ishkanian AS, Sykes J, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol 2014;15:1521-32. [Crossref] [PubMed]
- Blume-Jensen P, Berman DM, Rimm DL, et al. Development and clinical validation of an in situ biopsy-based multimarker assay for risk stratification in prostate cancer. Clin Cancer Res 2015;21:2591-600. [Crossref] [PubMed]
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028-38. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Dal Pra A, Lalonde E, Sykes J, et al. TMPRSS2-ERG status is not prognostic following prostate cancer radiotherapy: implications for fusion status and DSB repair. Clin Cancer Res 2013;19:5202-9. [Crossref] [PubMed]
- Locke JA, Zafarana G, Ishkanian AS, et al. NKX3.1 haploinsufficiency is prognostic for prostate cancer relapse following surgery or image-guided radiotherapy. Clin Cancer Res 2012;18:308-16. [Crossref] [PubMed]
- Ritter MA, Gilchrist KW, Voytovich M, et al. The role of p53 in radiation therapy outcomes for favorable-to-intermediate-risk prostate cancer. Int J Radiat Oncol Biol Phys 2002;53:574-80. [Crossref] [PubMed]


