Microarray screening for key genes and prognosis factors in interferon regulatory factor 1-silenced ovarian cancer SKOV-3 cells
Introduction
Ovarian cancer is a gynecological neoplastic disease and the fifth most common cause of cancer mortality in women (1). Survival of patients with ovarian cancer is reported to be highly related to the stage of cancer: 5-year survival rate for patients with early-stage cancer is 80–90%, whereas that for patients with advanced-stage disease is merely 25% (2).
Epithelial ovarian cancer, described as a “silent killer,” is the most common type of ovarian cancer (3). Approximately 90% of ovarian cancers affect the single-cell epithelial layer of the ovarian surface (4). However, timely adoption of preventive measures for ovarian cancer is difficult because of the lack of obvious symptoms during the early stage and dearth of effective early-diagnostic tools.
In the past, several studies have used ultrasound (5) and cancer antigen 125 (CA 125) (6) as the primary test for ovarian cancer. The CA 125 assay was used as first-line screening because of its relatively noninvasive nature during blood sampling. Serum CA 125 levels increased in 23–50% of surgical stage I and 90% of stage II ovarian carcinomas (7). However, rather than a prognostic or diagnostic marker, CA 125 level is used only for following the response or progression of the disease (8).
Recently, large-scale gene expression analysis has been used to screen differentially expressed genes (DEGs) in ovarian cancer (9), especially for identifying potential tumor markers of early-stage diagnosis and ensuring timely treatment (10). Transcription factors regulate the expression of tumor-associated genes (TAGs), which provides insights for research regarding the key genes in ovarian cancer (11). Interferon regulatory factor 1 (IRF-1), a member of the interferon regulatory transcription factor family, activates the transcription of interferons alpha and beta. It is also a tumor suppressor gene (TSG) that prevents oncogene-mediated malignant transformation (12). IRF-1 expression in tumors is an independent predictor of favorable clinical outcomes for ovarian cancer (13), and it is likely that gene expression could differ with IRF-1 silencing.
In this study, epithelial ovarian cancer SKOV-3 cells that were separately transfected with IRF-1 short hairpin ribonucleic acid (shRNA) and scrambled shRNA were used to analyze DEGs with IRF-1 silencing to understand the mechanism of ovarian cancer.
Methods
Microarray data
Microarray expression data was obtained from the platform data of GPL10558 (IlluminaHumanHT-12 V4.0 expression beadchip) from the Gene Expression Omnibus (GEO) database (accession number GSE38551; http://www.ncbi.nlm.nih.gov/geo/), which was deposited by Pavan et al. (12). The microarray included 12 samples [3 SKOV-3 samples transfected with scrambled shRNA, 3 with scrambled shRNA with cis-diamminedichloroplatinum (CDDP), 3 with IRF-1 shRNA, and 3 with IRF-1 shRNA with CDDP]. For analysis, we used 3 SKOV-3 samples transfected with IRF-1 shRNA and 3 with scrambled shRNA.
Data preprocessing and DEG analysis
Data preprocessing (background correction, quantile normalization, probe summarization) was performed using the robust multi-array average algorithm (14) in the Limma software; the t-test (15) was used to identify significantly expressed DEGs in SKOV-3 samples transfected with IRF-1 shRNA and those transfected with scrambled shRNA. A false discovery rate (FDR) <0.05 and an absolute value of log2FC (fold change) >1 were used as thresholds.
Gene ontology (GO) and pathway enrichment analysis for DEGs
GO analysis, including biological process (BP), molecular function (MF), and cellular component, is used for the unification of biology (16). The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database used to classify relevant gene sets into their respective pathways (17). In this study, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to identify significant GO categories in BPs and significant pathways with P<0.05.
Functional annotation for DEGs
Using the transcription factor data, we screened and annotated DEGs to determine whether they could regulate transcription. TSG (18) and TAG databases (19) were used for screening TSGs and oncogenes.
Construction of a PPI network
The Search Tool for the Retrieval of Interacting Genes (STRING) database can provide both experimental and predicted interaction information of proteins (20). In this study, STRING was used for protein-protein interaction (PPI) network analysis and confidence value (combined score) >0.4 was regarded as the threshold. Cytoscape was used to construct the PPI network, and highly connected nodes (hubs) (21) were obtained.
Subnetwork construction and enrichment analyses
To obtain further information regarding DEGs scored in the PPI network, a subnetwork was constructed using the BioNet software (22) in R with FDR =0.0001. GO and KEGG enrichment analyses were performed for DEG-encoded proteins in the subnetwork.
MiRNA-target regulating analysis
We performed microRNA (miRNA) prediction using WebGestalt GAST (23) (http://www.webgestalt.org/option.php), and conducted miRNA-target enrichment prediction for DEGs in the PPI network by overrepresentation enrichment analysis (ORA). The species was Hsapiens, the minimum number of enriched DEGs was 2, and results with P<0.05 were obtained.
Survival analysis
DEGs related to survival and prognoses were searched in The Cancer Genome Atlas (TCGA) database. The DEGs were then grouped by the median into high- and low-expression genes. Age, gender, and cancer stage were adjusted using the Cox model; P<0.05 was considered to be significant. Hazard ratios (HRs) of these DEGs were predicted for survival. High-expression DEGs with HR >1 and low-expression DEGs with HR <1 were screened, and Kaplan–Meier survival curves were drawn.
Results
DEG selection
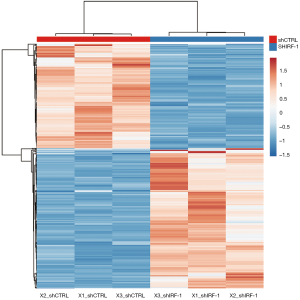
Results showed that 442 transcriptional factors were observed: 250 upregulated and 192 downregulated factors. In these 427 DEGs were obtained: 242 upregulated DEGs and 185 downregulated DEGs (Figure 1).
GO and KEGG enrichment analysis for DEGs
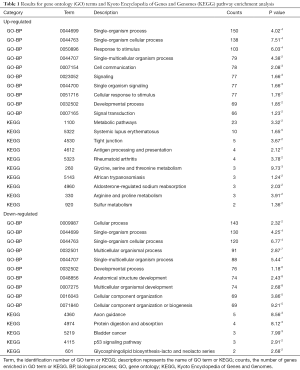
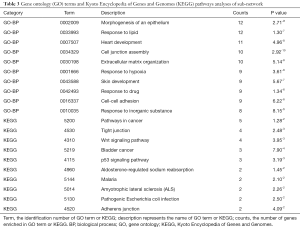
We performed GO and KEGG enrichment analyses using P<0.05 for the functional analysis of DEGs. Several GO categories were enriched among these DEGs, and Table 1 lists the top ten categories for up- and downregulated DEGs. Three categories of BPs enriched most DEGs with a count >100. In upregulated DEGs, the BPs were single-organism process, single-organism cellular process, and response to stimulus, and in downregulated DEGs, the BPs were cellular process, single-organism process, and single-organism cellular process. Table 1 also shows the pathways that were obtained by KEGG enrichment. In upregulated DEGs, a total of ten pathways with a count >2 were obtained, mainly metabolic pathways, systemic lupus erythematosus, and tight junctions. In downregulated genes, five pathways were obtained with a small count.
Full table
Functional annotation for DEGs
Results showed that eight transcriptional factors were upregulated and 11 downregulated (Table 2). In the upregulated factors, 26 genes were detected (2 oncogenes, 20 TSGs, and 4 genes with unknown functions), and in the downregulated factors, 19 genes were detected (3 oncogenes, 15 tumor genes, and 1 gene with whose unknown functions).

Full table
PPI network construction
We finally obtained 173 interaction pairs. In the PPI network (Figure 2), degrees of 14 proteins were >5: tumor necrosis factor (TNF), degree =28; CDH1, degree =20; matrix metallopeptidase 2 (MMP2), degree =13; collagen type I alpha 1 chain (COL1A1), degree =12; serpin family E-member 1 (SERPINE1), degree =12; MMP1, degree =9; gap junction protein alpha 1 (GJA1), degree =8; fibrillin 1 (FBN1), degree =8; Snail family transcriptional repressor 2 (SNAI2), degree =7; thrombospondin 1 (THBS1), degree =7; forkhead box O1 (FOXO1), degree =7; CCAAT/enhancer binding protein delta, degree =7; claudin 3 (CLDN3), degree =7; Dickkopf Wnt signaling pathway inhibitor 1 (DKK1), degree =7; integrin subunit beta 4 (ITGB4), degree =6; keratin 14 (KRT14), degree =6; tight junction protein 3 (TJP3), degree =6; KIT ligand (KITLG), degree =6; and MMP7, degree =6. The degrees of TNF and CDH1 were the top 2 nodes compared to other proteins.
Subnetwork analyses
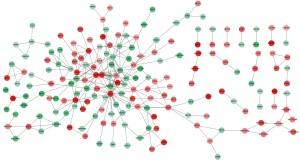
As shown in Figure 3, 48 nodes and 68 interaction pairs were included in the subnetwork. Among them, consistent with the results of the PPI network, the top 7 proteins with high degrees were TNF, CDH1, MMP2, GJA1, DKK1, THBS1, and SERPINE1. Table 3 lists the GO terms and KEGG pathway enrichment for the subnetwork. For GO terms, DEGs were mainly enriched in the BP of epithelium morphogenesis (e.g., CDH1) and response to lipids (e.g., TNF). KEGG analysis revealed that DEGs were mainly enriched in cancer pathways (e.g., CDH1 enriched), tight junctions, Wnt signaling pathway (e.g., DKK1 enriched), bladder cancer (e.g., CDH1, THBS1 enriched), and p53 signaling pathway (e.g., THBS1, SERPINE1 enriched).
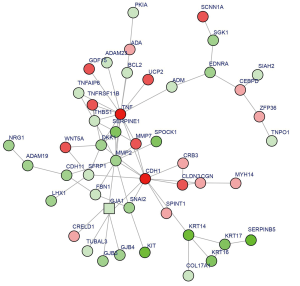
Full table
MiRNA-target-regulating analysis
Table 4 present results for miRNA-target-regulating analysis. In this study, we identified 15 miRNAs targeting DEGs. Among these miRNAs, miR-30A-5p, miR-30C, miR-30D, miR-30B, and miR-30E-5p targeted the TGTTTAC motif contained in 25 DEGs, including FRMD6, FAM43A, EDNRA, EPHB2, and KHNYN. MiR-498 targeted the GCTTGAA motif in nine DEGs, including COL1A1, DUSP4, FLRT2, HBP1, and GJA1. MiR-492 was predicted to target five DEGs: TSKU, C17orf58, STC1, ZFP36, and SH3PXD2A. MiR-489 targeted six DEGs: ADAMTS5, PRSS23, FBN1, NRG1, SGK1, and WNT5A. Both miR-27A and miR-27B were predicted to target 17 DEGs, including CDH11, E2F7, DCP2, EDNRA, EPHB2, and FLRT2.
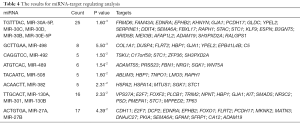
Full table
Survival analysis
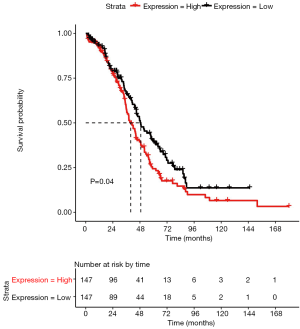
We isolated ovarian cancer prognosis-associated data from TCGA database and identified survival-correlated DEGs according to the selection criteria. As a result, 38 prognosis-associated genes (e.g., CDH1) were obtained. Specifically, CDH1, which was upregulated in IRF-1-silenced SKOV3 cells, was predicted to be negatively correlated with survival in patients with ovarian cancer (Figure 4).
Discussion
In this study, we found 427 DEGs (242 upregulated and 185 downregulated) and their function categories that altered in IRF-1-silenced SKOV-3 cell samples. Pathway enrichment analyses for all DEGs showed that genes were mainly enriched in metabolic pathways and systemic lupus erythematosus. TNF and CDH1 had a higher degree than others in both the PPI network and the subnetwork. Furthermore, GO and KEGG enrichment analyses for the subnetwork showed that CDH1 was enriched in the BP of epithelium morphogenesis and cancer pathways, and TNF was enriched in response to lipids. Besides, survival analysis showed that CDH1 was associated with ovarian cancer prognosis.
A previous study reported the significant role of cancer cell metabolism pathways in colorectal carcinomas (24), which suggested that cancer cells share common enzyme or transporter activities, suggestive of anaerobic metabolism with high ability for lactate extrusion and glucose absorption. Otherwise, it is difficult for the tumor to survive and grow. Systemic lupus erythematosus is a prototypical autoimmune disease characterized by the production of immunoglobulin G (IgG) autoantibodies that are specific for self-antigens. Bernatsky et al. (25) proved the association between systemic lupus erythematosus and cancer and indicated that certain cancers occur more frequently in patients with systemic lupus erythematosus than in those without. Our results of pathway enrichment analyses of all DEGs showed that the genes were mainly enriched in metabolic pathways and systemic lupus erythematosus. Thus, metabolic pathways and systemic lupus erythematosus may be involved in ovarian cancer.
TNF encodes a multifunctional pro-inflammatory cytokine belonging to the TNF superfamily from the CDH superfamily, and its high expression in ovarian cancer cells indicates its importance (26). Son et al. suggested that targeting pro-inflammatory chemokines induced by epidermal growth factor (EGF) or TNF, including CCL20, CXCL1-3, and CXCL8, may be a potential treatment for ovarian cancer with many epidermal growth factor receptor and TNF activation patterns (27).Tania et al. reported that lipid metabolism is related to ovarian cancer risk, and targeting enzymes of lysophosphatidic acid metabolism might be useful for further cancer therapy (28). In our study, TNF was significantly enriched in the BP of response to lipids and might play a significant role in ovarian cancer via this response.
CDH1, a classic cadherin, is expressed predominantly on epithelial cell surface and plays a key role in the maintenance and establishment of normal tissue architecture (29). It encodes a calcium-dependent cell-cell adhesion glycoprotein formed by five extracellular cadherin repeats. Studies have reported that transfection of human cancer cell lines with E-cadherin complementary deoxyribonucleic acid (cDNA) can decrease their invasiveness (30). This may explain why CDH1 expression in epithelial ovarian cancer cells is upregulated with IRF-1 silencing. Combined detection of serum human epididymis protein 4 (HE4) and CDH1 gene methylation levels could help differentiate ovarian endometriosis cysts from ovarian cancer during diagnosis (31). A previous meta-analysis indicated that CDH1 promoter methylation could be a potential biomarker in ovarian cancer risk prediction (32). In our study, CDH1 was enriched in the BP of epithelial morphogenesis and cancer pathways. In addition, survival analysis showed that CDH1 is associated ovarian cancer prognosis. Therefore, our study further confirmed that CDH1 is an important prognosis factor for ovarian cancer, using bioinformatics analysis. CDH1 may be involved in this cancer mainly via the BP of epithelial morphogenesis and cancer pathways.
Previous studies have demonstrated that the miR-30 family plays a critical role in cancer pathogenesis. Ouzounova et al. documented that the miR-30 family regulates nonattachment growth of breast cancer cells (33). Another study reported that miR-30-5p acts as a tumor suppressor to regulate multiple myeloma pathogenesis by targeting the Wnt/β-Catenin/BCL-9 pathway (34). In addition, miR-30 also acts as a tumor suppressor to inhibit epithelial-mesenchymal transition (EMT) in prostate cancer via the EGF/Src tyrosine kinase pathway (35). In ovarian cancer, miR-30a overexpression could highly reduce the expression of proliferating cell nuclear antigen (PCNA), a common marker for proliferation, in human ovarian granulosa cells (36). Ye et al. reported that miR-30D suppresses transforming growth factor β1 (TGF-β1)-induced EMT by targeting Snail (a major determinant of ovarian cancer invasiveness at the transcription level) in ovarian cancer (37). Taken together, the miR-30 family may act as a tumor suppressor in ovarian cancer. In our study, the miR-30 family, including miR-30A, miR-30B, miR-30C, miR-30D, and miR-30E, was predicted to target several DEGs identified, such as ADAM19, FRMD6, and STC1. Therefore, it is important to further reveal the regulatory mechanism of the miR-30 family in ovarian cancer.
In conclusion, TNF, CDH1, and the miR-30 family may play significant roles in ovarian cancer. CDH1 is a vital prognosis factor for ovarian cancer and might be involved via the BP of epithelium morphogenesis and cancer pathways. TNF plays a vital role via the BP of response to lipids, and the MiR-30 family may serve as a tumor suppressor in ovarian cancer pathogenesis. One of the limitations of this study is the lack of verification; thus, further verification experiments are required.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (81771594, 81671474 and 81501267), Natural Science Foundation of Guangdong Province (2017A030313460), the Major Project of the Department of Science and Technology of Guangdong Province (2014B020213001 and 2013B022000005), the Major Project of the Department of Science and Technology of Guangzhou (2014Y2-00059, 201604020012 and 201704020108).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.10). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schwede M, Spentzos D, Bentink S, et al. Stem Cell-Like Gene Expression in Ovarian Cancer Predicts Type II Subtype and Prognosis. PLoS One 2013;8:e57799 [Crossref] [PubMed]
- Colombo N, Van Gorp T, Parma G, et al. Ovarian cancer. Crit Rev Oncol Hematol 2006;60:159-79. [Crossref] [PubMed]
- Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer 2005;5:355-66. [Crossref] [PubMed]
- Auersperg N, Wong AS, Choi KC, et al. Ovarian Surface Epithelium: Biology, Endocrinology, and Pathology. Endocr Rev 2001;22:255-88. [PubMed]
- Sato S, Yokoyama Y, Sakamoto T, et al. Usefulness of mass screening for ovarian carcinoma using transvaginal ultrasonography. Cancer 2000;89:582-8. [Crossref] [PubMed]
- Jacobs IJ, Skates S, Davies AP, et al. Risk of diagnosis of ovarian cancer after raised serum CA 125 concentration: a prospective cohort study. BMJ 1996;313:1355-8. [Crossref] [PubMed]
- Zurawski VR Jr, Orjaseter H, Andersen A, et al. Elevated serum CA 125 levels prior to diagnosis of ovarian neoplasia: relevance for early detection of ovarian cancer. Int J Cancer 1988;42:677-80. [Crossref] [PubMed]
- Meyer T, Rustin GJ. Role of tumour markers in monitoring epithelial ovarian cancer. Br J Cancer 2000;82:1535-8. [PubMed]
- Skubitz AP, Pambuccian SE, Argenta PA, et al. Differential gene expression identifies subgroups of ovarian carcinoma. Transl Res 2006;148:223-48. [Crossref] [PubMed]
- Jiang X, Zhu T, Yang J, et al. Identification of novel epithelial ovarian cancer biomarkers by cross-laboratory microarray analysis. J Huazhong Univ Sci Technolog Med Sci 2010;30:354-9. [Crossref] [PubMed]
- Ying H, Lv J, Ying T, et al. MicroRNA and transcription factor mediated regulatory network for ovarian cancer. Tumour Biol 2013;34:3219-25. [Crossref] [PubMed]
- Pavan S, Olivero M, Corà D, et al. IRF-1 expression is induced by cisplatin in ovarian cancer cells and limits drug effectiveness. Eur J Cancer 2013;49:964-73. [Crossref] [PubMed]
- Zeimet AG, Reimer D, Wolf D, et al. Intratumoral interferon regulatory factor (IRF)‐1 but not IRF‐2 is of relevance in predicting patient outcome in ovarian cancer. Int J Cancer 2009;124:2353-60. [Crossref] [PubMed]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249-64. [Crossref] [PubMed]
- Smyth GK, Ritchie M, Thorne N, et al. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, et al. editors. Bioinformatics and computational biology solutions using R and bioconductor. Berlin: Springer, 2005:397-420.
- Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc 2009;3:S10. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res 2013;41:D970-6. [Crossref] [PubMed]
- Chen JS, Hung WS, Chan HH, et al. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics 2013;29:420-7. [Crossref] [PubMed]
- von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258-61. [Crossref] [PubMed]
- He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet 2006;2:e88 [Crossref] [PubMed]
- Beisser D, Klau GW, Dandekar T, et al. BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics 2010;26:1129-30. [Crossref] [PubMed]
- Wang J, Duncan D, Shi Z, et al. WEB-based gene set analysis toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41:W77-83 [Crossref] [PubMed]
- Koukourakis MI, Giatromanolaki A, Harris AL, et al. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res 2006;66:632-7. [Crossref] [PubMed]
- Bernatsky S, Boivin JF, Joseph L, et al. An international cohort study of cancer in systemic lupus erythematosus. Arthritis Rheum 2005;52:1481-90. [Crossref] [PubMed]
- Macciò A, Madeddu C. Inflammation and ovarian cancer. Cytokine 2012;58:133-47. [Crossref] [PubMed]
- Son DS, Kabir SM, Dong Y, et al. Characteristics of chemokine signatures elicited by EGF and TNF in ovarian cancer cells. J Inflamm (Lond) 2013;10:25. [Crossref] [PubMed]
- Tania M, Khan MA, Song Y. Association of lipid metabolism with ovarian cancer. Curr Oncol 2010;17:6-11. [PubMed]
- Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta 1994;1198:11-26. [PubMed]
- Frixen UH, Behrens J, Sachs M, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol 1991;113:173-85. [Crossref] [PubMed]
- Sun B, Zhang X. Value of abnormal methylation of CDH1 gene and the detection of serum HE4 in the identification of ovarian cancer and ovarian endometriosis cyst. Hainan Med J 2015;26:3023-5.
- Wang Q, Wang B, Zhang YM, et al. The association between CDH1 promoter methylation and patients with ovarian cancer: a systematic meta-analysis. J Ovarian Res 2016;9:23. [Crossref] [PubMed]
- Ouzounova M, Vuong T, Ancey PB, et al. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics 2013;14:139. [Crossref] [PubMed]
- Zhao JJ, Lin J, Zhu D, et al. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res 2014;74:1801-13. [Crossref] [PubMed]
- Kao CJ, Martiniez A, Shi XB, et al. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014;33:2495-503. [Crossref] [PubMed]
- Sirotkin AV, Lauková M, Ovcharenko D, et al. Identification of MicroRNAs controlling human ovarian cell proliferation and apoptosis. J Cell Physiol 2010;223:49-56. [PubMed]
- Ye Z, Zhao L, Li J, et al. miR-30d Blocked Transforming Growth Factor β1-Induced Epithelial-Mesenchymal Transition by Targeting Snail in Ovarian Cancer Cells. Int J Gynecol Cancer 2015;25:1574-81. [Crossref] [PubMed]