Effects of three common polymorphisms in microRNAs on lung cancer risk: a meta-analysis
Introduction
Lung cancer is the most common malignant cancer, which causes approximately 1.59 million cases deaths, with less than 15% of the 5 years survival rate according to World Health Organization (WHO) (1,2). For Chinese, lung cancer is the second common malignant tumor in female and the most frequent malignant tumor in male. Tumorigenesis is a very complex process (3). And the etiology of lung cancer is also strongly affected by genetic or environmental factors and the interaction of gene-environment. Molecular epidemiological studies also indicated that a large number of genetic variants might be associated with the risk of lung cancer (4). The GWAS studies indicated that there were many disease-associated loci in non-coding RNAs (5,6). MicroRNAs played pivotal roles in non-coding RNAs and were associated with cancer cell proliferation, invasion and cell cycle regulation. Many studies shown that miRNAs could influence the initiation of cancer or the progression of malignant tumor (7). Thus, the abnormal expressions of miRNAs may be the cause of lung cancer (8). Single nucleotide polymorphisms (SNPs) in microRNAs could affect the cancer risk by changing the expressions of miRNAs (9). In the past, many studies shown that the three common polymorphisms [miR-146a (rs2910164), miR-196a2 (rs11614913) and miR-499 (rs3746444)] could influence the susceptibility of lung cancer, but the results were contradictory. The common variants [minor allele frequency (MAF) >0.05] were concerned in previous studies, such as rs2910164, rs11614913 and rs3746444 (10).
MiR-146a (rs2910164) is exits on chromosome fifth LOC285628, and the mature miR-146a is located in the second exon (11,12). MiR-146a played an important role in a majority of disease (13) and it also can influence the occurrence and development of tumor by disturbing cell invasion and migration (14,15). Many studies have conducted to estimate the potential association between miR-146a and lung cancer in humans. According to the search strategy, we found six studies to analysis the association between miR-146a and lung cancer (16-21).
MiR-196a-2 (rs11614913) located the mature miRNA complementary region of pre-miR-196a-2, and a large number of studies have shown that the SNP was associated with the risk of lung cancer by influencing the expression and maturation of miRNAs (19-24).
MiR-499 gene is also exiting in miRNA mature region. MiR-499 plays an important role in the regulation of cell differentiation. Some studies have indicated that miR-499 (rs3746444) polymorphism was associated with the risk of lung cancer (7,19,20) (Qiu F, 2012, unpublished data).
The aim of this meta-analysis was to achieve a combined risk estimate including most recently published studies before Apr 2017. And we were evaluated the relationship between these three polymorphisms (miR-146a, miR-196a2, and miR-499) with lung cancer risk.
Methods
Data collection
We conducted a comprehensive and systematic search to analyze the association between these three common polymorphisms and lung cancer in PubMed, Wanfang, VIP, CNKI database. Last search was conducted on April 2017. We used the subject headings and keywords such as, miRNAs, cancer, tumor, gene, polymorphism, variation, miR-499 (rs3746444), miR-146a (rs2910164), miR-196a2 (rs11614913) and supplemented by literature tracing methods to collect relevant studies.
Inclusion criteria
The Inclusion criteria were: (I) published all over the world about miR-146a (rs2910164), miR-196a2 (rs11614913) and miR-499 (rs3746444) polymorphisms and lung cancer risk from case-control study; (II) providing the number of the case group and the control group; (III) obtaining the full text of the literature; (IV) providing enough data to calculate the statistical index of odds ratio (OR) 95% confidence interval (CI); (V) the similar method and assumption of each research; (VI) getting the consensus for each research from two inspectors.
Exclusion criteria
The exclusion criteria were: (I) duplicated data; (II) meeting, case report and literature review; (III) involving gene expression, meta-analysis, cell lines; (IV) the Newcastle-Ottawa scale (NOS) quality assessment less than five stars (25).
Information extraction
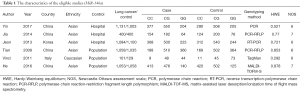
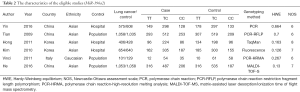
Two investigators independently extracted data. The following data were extracted: the first author’s name, published time, country, race, control source, number of cases and controls, the number of cases and controls of each genotype, the detection method of miRNA, Hardy-Weinberg equilibrium (HWE), NOS (Tables 1-3). If difference was existed after data collection, the third author needs to ensure the data. HWE was used to evaluate the gene frequency and genotype frequency by the goodness of fit χ2 test or Chi-square test in each study control groups. The disequilibrium was exited, when P<0.05. We assessed the association between the three SNPs and lung cancer risk by using ORs with 95% CIs (P<0.05). We analyzed the pooled ORs using five different genetic models: dominant model, homozygote comparison, recessive model, allelic comparison and heterozygote comparison, respectively. Furthermore, subgroup analyses carried out by race and control source. Heterogeneity between the eligible studies was assessed using Chi-square-based t-test. When I2≤50%, heterogeneity is not apparent. We can choose fixed effects model; whereas I2>50%, we think the heterogeneity exiting between studies, we should choose the random effect model (26,27). Further, to test the stability of the results, an accurate sensitivity analysis was needed by omitting one by one. We used quantitative method to determine publication bias in our meta-analysis (Egger’s test) (28,29). All analyses results were performed by using Stata software.
Full table
Full table
Full table
Results
The selection of eligible studies
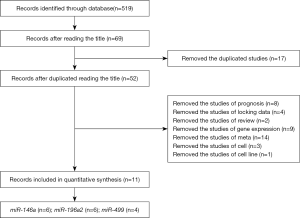
According to the search strategy, we got 519 articles from PubMed database, CNKI, Wanfang database and Chinese biomedical literature database. We removed 450 records by primary screening of titles. Figure 1 shows the screening process of studies. We excluded 58 articles (17 studies were duplicate records, 8 studies were prognosis, 3 studies were involved in cells, 9 studies were gene expression, 14 studies were meta-analyses, 2 studies were review, 4 studies did not have the data which we needed, 1 study was cell line). Finally, there were 11 studies in our meta-analysis with 9,231 lung cancer cases and 9,280 controls. Six studies about miR-146a (rs2910164) polymorphism were included. Six studies about miR-196a2 (rs11614913) polymorphism were analyzed. Four studies (5 data) about miR-499 (rs3746444) were entered.
The results of quantitative analysis
MiR-146a
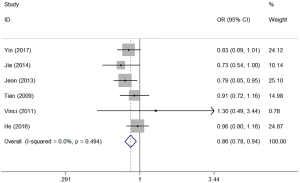
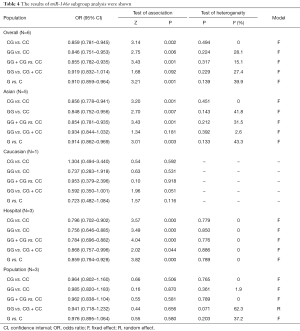
In this study, we set five different genetic models of miR-146a (CG vs. CC, GG vs. CC, GG + CG vs. CC, GG vs. CG + CC, G vs. C). Table 4 showed all the detailed results. The analysis revealed that miR-146a polymorphism was significantly associated with the risk of lung cancer (CG vs. CC: OR =0.859, 95% CI: 0.781–0.945, P=0.002; GG vs. CC: OR =0.846, 95% CI: 0.751–0.953, P=0.006; GG + CG vs. CC: OR =0.855, 95% CI: 0.782–0.935, P=0.001; G vs. C: OR =0.910, 95% CI: 0.859–0.964, P=0.001). In the subgroup analysis by source of controls, statistically decreased lung cancer risk was found in hospital- based groups (Table 4). No significant association between miR-146a and lung cancer risk was found in the population-based groups. Stratified analysis by ethnicity, the miR-146 a polymorphism was significantly associated with lung cancer risk among Asians (Table 4). Figure 2 shows the forest plot of miR-146a (CG vs. CC).
Full table
MiR-196a2
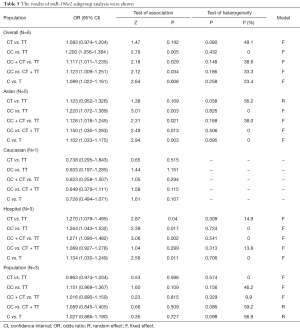
In this study, we set the heterozygote comparison of miR-196a2 (CT vs. TT), homozygote comparison (CC vs. TT), dominant model (CC + CT vs. TT), recessive model (CC vs. CT + TT), and allelic comparison (C vs. T). The analysis revealed that miR-196a2 polymorphism was significantly associated with increased lung cancer risk (CC vs. TT: OR =1.200, 95% CI: 1.056–1.364, P=0.005; CC + CT vs. TT: OR =1.117, 95% CI: 1.011–1.235, P=0.029; CC vs. CT + TT: OR =1.123, 95% CI: 1.009–1.251, P=0.034; C vs. T: OR =1.089, 95% CI: 1.022–1.161, P=0.008). In the stratified analysis by ethnicity, the miR-196a2 polymorphism was significantly associated with lung cancer risk in Asians (Table 5). In the subgroup analysis by source of controls, we found a statistically association between miR-196a2 and the lung cancer risk among hospital-based groups (Table 5). The forest plot of miR-196a2 (CC vs. TT) was shown in Figure 3.
Full table
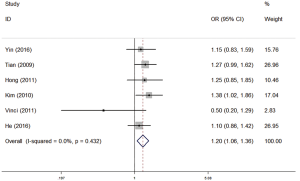
MiR-499
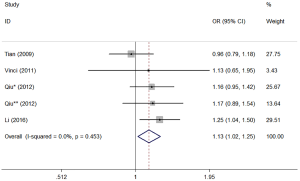
Table 6 listed the results of each genetic model. The analysis showed that miR-499 polymorphism might have association with lung cancer risk in the five different genetic models (AG vs. AA: OR =1.131, 95% CI: 1.022–1.252, P=0.018; GG vs. AA: OR =1.702, 95% CI: 1.093–2.650, P=0.019; AG + GG vs. AA: OR =1.207, 95% CI: 1.042–1.398, P=0.012; GG vs. AG + AA: OR =1.640, 95% CI: 1.066–2.523, P=0.024; G vs. A: OR =1.226, 95% CI: 1.026–1.464, P=0.025). Results of the stratified analysis by ethnicity suggested that the significant effect for miR-499 polymorphism was observed in the risk of lung cancer in among Asian (Table 6). Figure 4 shows the forest plot of miR-499 (AG vs. AA).
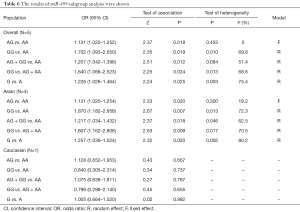
Full table
The analysis of sensitivity
We analyzed the sensitivity analysis by excluding study one by one. The analysis of sensitivity was suggesting that no obviously effects were exited from each article. That was say, our results were stable.
Publication bias analysis
We performed quantitative analysis by Egger’s test. We found P>0.05 and 95% CI including 1 in this study, suggesting no meaningful bias in this meta-analysis.
Discussion
In the past years, majority studies have shown that miRNAs play vital roles in cancer risks (30-32). And the newly miRNAs have enjoyed a high level of concern in medical science. MiR-219-1 (rs213210 and rs107822) might be associated with lung cancer risk (33). A significant association between the miR-608 (rs4919510) polymorphism and lung cancer risk was also observed (34). Rs12740674 in miR-1262 was significantly related with increased risk of lung cancer (35). Many studies have proved that miR-146a (rs2910164), miR-196a2 (rs11614913) and miR-499 (rs3746444) were related to the incidence of lung cancer. Compared with other SNPs, these three common polymorphisms in microRNAs were more popular and hot. MiR-146a (2910164), miR-196a2 (rs11614913) and miR-499 (rs3746444) polymorphism may have different function in the variety of tumors (36-38).The down-regulation of miR-146a contributed to COX-2 expression levels in lung cancer cells, including A549 cells (39). By influencing the expression and maturation of miRNAs, miR-196a2 was associated with the risk of lung cancer (19). MiR-196a2 was related with a wide variety of malignancies, including non-small-cell lung cancer (40). MiR-499 might play an important role for early detection non-small cell lung cancer (NSCLC) (41). For lung cancer, previous meta-analysis did not get the same results about miR-146a and miR-196a2 in some aspects. There was a meta-analysis found that miR-146a was not associated with lung cancer by Xu et al. (42). Xu et al. found that miR-196a2 was associated with lung cancer risk. Ren et al. (43) indicated that miR-146a might have significant association with lung cancer risk, particularly in Asians and the control from hospital-base. For lung cancer, miR-196a2 was a dangerous factor in Asians and the control from population-base.
It was worth noting that there were several problems in the above meta-analyses. As we can see, the association of miR-146a, miR-196a2 and miR-499 with lung cancer susceptibility could not be clear observed by previous meta-analysis. They got the different results might be limited the number of articles. Recently, we found more new researches in these fields and the sample size was large. Also, our own laboratory has been engaged in this research. Therefore, an update meta-analysis was needed to show the clear association between three common polymorphisms and lung cancer risk. Comparing with the previous meta-analysis, this meta-analysis including 9,231 cases and 9,280 controls combined previously published articles and an academic dissertation was conducted for the aim of estimating the real relation between three common polymorphisms and the risk of lung cancer. For all we know, this is the largest meta-analysis to evaluate the relationship between the three common polymorphisms in miRNAs and lung cancer risk.
In the overall analysis, results implied that miR-146a (rs2910164) had a significant correlation with the risk of lung cancer. And compared with homozygous CC, homozygous GG might be a protective factor for lung cancer. Compared with allele C, allele G was found to be associated with decreased lung cancer risk. Subgroup analyses based on ethnicity, results indicated that significantly affected lung cancer risk was found for miR-146a in Asians when subgroup analysis by sources of controls, it performed that miR-146a (rs2910164) had obvious correlation with lung cancer risk in the hospital-based study but not in population-based study. And compared with homozygous CC, homozygous GG might be associated with decreased the risk of lung cancer. Compared with allele C, allele G could decrease the risk of lung cancer. In the overall analysis, results presented that miR-196a2 (rs11614913) had obvious correlation with the risk of lung cancer. Homozygous CC might be the risk factor for lung cancer compared with homozygous TT. By observing recessive model, we could find the homozygous CC increased the risk of lung cancer. The subgroup analysis by race, individuals with homozygous CC had the higher risk of lung cancer in Asian population compared with homozygous TT. When subgroup analyzed by sources of controls, homozygous CC/heterozygous CT could be associated with increased lung cancer risk compared with homozygous TT in the hospital-based study. In the overall analysis, results presented that miR-499 had correlation with the risk of lung cancer. MiR-499 might be related with increased lung cancer risk in five genetic models. The subgroup analyzed by race, the results of five genetic models showed the significant association between miR-499 and the risk of lung cancer.
Although our research efforts are sufficient to implement a comprehensive analysis, there are still some limitations. Firstly, the different genetic backgrounds and living environments were exited in these conducted studies (44,45). Then, these case control studies only included Asian (Chinese and Korean populations) and Caucasian (Italy population). So the results could not be used as a model for other countries. Due to the limited number of Caucasians study, the results could not be used as a model for Caucasians. Secondly, publication bias might be existed, but the results of publication bias did not have statistical significance. Third, since the original studies did not have the smoking status, it was not possible to conduct subgroup analysis on smoking. Finally, we only analysis the published studies but not include unpublished articles.
Conclusions
In conclusion, this meta-analysis provides evidence that miR-146a (rs2910164), miR-499 (rs3746444) and miR-196a2 (rs11614913) polymorphisms might contribute to associating with lung cancer risk. As we know, this is the first time to get the significant relationship between miR-499 and lung cancer risk. Also, the result needs to be confirmed by a series of further experiments.
In this meta-analysis, miR-146a (rs2910164), miR-196a2 (rs11614913) and miR-499 (rs3746444) may be associated with the risk of lung cancer.
Acknowledgments
Funding: This study was supported by Natural Science Foundation of Liaoning Province (No. 201602870).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.32). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parkin DM, Bray F, Ferlay J, et al. Estimating the world cancer burden: Globocan 2000. Int J Cancer 2001;94:153-6. [Crossref] [PubMed]
- Rong B, Yang S, Li W, et al. Systematic review and meta-analysis of Endostar (rh-endostatin) combined with chemotherapy versus chemotherapy alone for treating advanced non-small cell lung cancer. World J Surg Oncol 2012;10:170. [Crossref] [PubMed]
- Lou J, Gong J, Ke J, et al. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis 2017;38:177-83. [PubMed]
- Yuan Z, Zeng X, Yang D, et al. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PLoS One 2013;8:e61047 [Crossref] [PubMed]
- Goulart LF, Bettella F, Sonderby IE, et al. MicroRNAs enrichment in GWAS of complex human phenotypes. BMC Genomics 2015;16:304. [Crossref] [PubMed]
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009;106:9362-7. [Crossref] [PubMed]
- Li D, Zhu G, Di H, et al. Associations between genetic variants located in mature microRNAs and risk of lung cancer. Oncotarget 2016;7:41715-24. [PubMed]
- Zhu Y, Li T, Chen G, et al. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer 2017;114:6-11. [Crossref] [PubMed]
- Pandey N, Pal S, Sharma LK, et al. SNP rs16969968 as a Strong Predictor of Nicotine Dependence and Lung Cancer Risk in a North Indian Population. Asian Pac J Cancer Prev 2017;18:3073-9. [PubMed]
- Li J, Zou L, Zhou Y, et al. A low-frequency variant in SMAD7 modulates TGF-beta signaling and confers risk for colorectal cancer in Chinese population. Mol Carcinog 2017;56:1798-807. [Crossref] [PubMed]
- Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum 2008;58:1284-92. [Crossref] [PubMed]
- Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006;103:12481-6. [Crossref] [PubMed]
- Saba R, Sorensen DL, Booth SA. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front Immunol 2014;5:578. [Crossref] [PubMed]
- Labbaye C, Testa U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol 2012;5:13. [Crossref] [PubMed]
- Xiang Z, Song J, Zhuo X, et al. MiR-146a rs2910164 polymorphism and head and neck carcinoma risk: a meta-analysis based on 10 case-control studies. Oncotarget 2017;8:1226-33. [Crossref] [PubMed]
- Yin Z, Cui Z, Ren Y, et al. MiR-146a polymorphism correlates with lung cancer risk in Chinese nonsmoking females. Oncotarget 2017;8:2275-83. [PubMed]
- Jia Y, Zang A, Shang Y, et al. MicroRNA-146a rs2910164 polymorphism is associated with susceptibility to non-small cell lung cancer in the Chinese population. Med Oncol 2014;31:194. [Crossref] [PubMed]
- Jeon HS, Lee YH, Lee SY, et al. A common polymorphism in pre-microRNA-146a is associated with lung cancer risk in a Korean population. Gene 2014;534:66-71. [Crossref] [PubMed]
- Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev 2009;18:1183-7. [Crossref] [PubMed]
- Vinci S, Gelmini S, Pratesi N, et al. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med 2011;49:2073-80. [Crossref] [PubMed]
- He F, Lin J, Yu T, et al. Interaction research on smoking and microRNA genes SNP related to lung cancer in Fujian Han population. Zhonghua Yu Fang Yi Xue Za Zhi 2016;50:168-74. [PubMed]
- Hong YS, Kang HJ, Kwak JY, et al. Association between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. J Prev Med Public Health 2011;44:125-30. [Crossref] [PubMed]
- Kim MJ, Yoo SS, Choi YY, et al. A functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean population. Lung Cancer 2010;69:127-9. [Crossref] [PubMed]
- Yin Z, Cui Z, Ren Y, et al. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer 2016;94:15-21. [Crossref] [PubMed]
- Cota GF, de Sousa MR, Fereguetti TO, et al. Efficacy of anti-leishmania therapy in visceral leishmaniasis among HIV infected patients: a systematic review with indirect comparison. PLoS Negl Trop Dis 2013;7:e2195 [Crossref] [PubMed]
- Xu Y, Gu L, Pan Y, et al. Different effects of three polymorphisms in MicroRNAs on cancer risk in Asian population: evidence from published literatures. PLoS One 2013;8:e65123 [Crossref] [PubMed]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820-6. [Crossref] [PubMed]
- Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [Crossref] [PubMed]
- Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol 2001;54:1046-55. [Crossref] [PubMed]
- Zhang L, Shan X, Wang J, et al. A three-microRNA signature for lung squamous cell carcinoma diagnosis in Chinese male patients. Oncotarget 2017;8:86897-907. [PubMed]
- Liu J, Ni S. Association between genetic polymorphisms in the promoters of let-7 and risk of cervical squamous cell carcinoma. Gene 2018;642:256-60. [Crossref] [PubMed]
- Sun R, Liang Y, Yuan F, et al. Functional polymorphisms in the promoter region of miR-17-92 cluster are associated with a decreased risk of colorectal cancer. Oncotarget 2017;8:82531-40. [PubMed]
- Zheng C, Li X, Xia L, et al. Polymorphisms of pri-miR-219-1 are associated with the susceptibility and prognosis of non-small cell lung cancer in a Northeast Chinese population. Oncotarget 2017;8:56533-41. [PubMed]
- Wu S, Yuan W, Shen Y, et al. The miR-608 rs4919510 polymorphism may modify cancer susceptibility based on type. Tumour Biol 2017;39:1010428317703819 [Crossref] [PubMed]
- Xie K, Chen M, Zhu M, et al. A polymorphism in miR-1262 regulatory region confers the risk of lung cancer in Chinese population. Int J Cancer 2017;141:958-66. [Crossref] [PubMed]
- Araújo R, Santos JM, Fernandes M, et al. Expression profile of microRNA-146a along HPV-induced multistep carcinogenesis: a study in HPV16 transgenic mice. J Cancer Res Clin Oncol 2018;144:241-8. [Crossref] [PubMed]
- Bodal VK, Sangwan S, Bal MS, et al. Association between Microrna 146a and Microrna 196a2 Genes Polymorphism and Breast Cancer Risk in North Indian Women. Asian Pac J Cancer Prev 2017;18:2345-8. [PubMed]
- Zhang H, Zhang Y, Yan W, et al. Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: a meta-analysis. Oncotarget 2017;8:393-407. [PubMed]
- Cornett AL, Lutz CS. Regulation of COX-2 expression by miR-146a in lung cancer cells. Rna 2014;20:1419-30. [Crossref] [PubMed]
- Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 2008;118:2600-8. [PubMed]
- Li M, Zhang Q, Wu L, et al. Serum miR-499 as a novel diagnostic and prognostic biomarker in non-small cell lung cancer. Oncol Rep 2014;31:1961-7. [Crossref] [PubMed]
- Xu L, Tang W. The associations of nucleotide polymorphisms in mir-196a2, mir-146a, mir-149 with lung cancer risk. Cancer Biomark 2015;15:57-63. [Crossref] [PubMed]
- Ren YG, Zhou XM, Cui ZG, et al. Effects of common polymorphisms in miR-146a and miR-196a2 on lung cancer susceptibility: a meta-analysis. J Thorac Dis 2016;8:1297-305. [Crossref] [PubMed]
- Wang P, Xie S, Cui A, et al. miR-196a2 polymorphisms and susceptibility to cancer: A meta-analysis involving 24,697 subjects. Exp Ther Med 2012;3:324-30. [Crossref] [PubMed]
- Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med 2002;4:45-61. [Crossref] [PubMed]