
The microbiota impact: bacteria shaping immunity, disease and response to therapy
Microbiota is an essential community of microorganisms that colonize from birth in different areas of the human body predominantly gut, oral and nasal mucosa, vaginal tract, etc. Is a dynamic population of over a trillion of microbes that include bacteria belonging to the different families, viruses and fungi that interact one with the other, with the local habitat and environment. The microbiome is the incredible number of genes that can be extrapolated by this complex community of cells being 100-fold larger than the whole genome. This fascinating “forgotten organ” is known to influence the training of the immune system thus affecting several physiological functions of the body and several diseases. Moreover, it contributes to the establishment of mechanisms for pathogen recognition and protection. Several factors are known to influence the variable part of the microbiome such as host lifestyle, environment, diet, genotype, pathobiology, inflammation, etc. Parameters associated with a healthy and fit microbiome are essentially the high diversity and a resilient feature, i.e., the capacity to cope with a physiological stress. An unbalanced scenario together with concomitant factors such as an inflammation process and/or presence of pathogens can cause complex diseases. Microbiota dysbiosis is thought to cause then several disorders such as inflammatory bowel diseases but also other systemic diseases like diabetes and interestingly central nervous system pathologies (1). In the last years the advent of molecular techniques such as next generation sequencing (NGS) have given a relevant impulse in the characterization of microbiome. Methodologies analyzing bacterial 16S rRNA and the more sophisticated DNA shotgun sequencing has allowed to perform targeted-metagenomic and full sequencing analyses of the microbial communities, thus defining bacterial phyla, families, genera down to species details, hence providing metataxonomic maps of microbiota. We now can define the portraits of bacteria colonizing the gut during lifetime and alterations of the composition linked to several diseases both in term of fecal and mucosal microbiota. Specific parameters of microbiota profiles, such as ecological biodiversity, differences in the relative abundances of taxa as well as the cataloguing of microbial sequences into operational taxonomic units (OUT) and their correlations can now identify microbial signatures associated to inflammation or other intestinal and extra-intestinal features and specific microbial biomarkers of disease.
In particular since microbiota is strictly associated with immunity and the development of a healthy immune system, it was not surprising the finding that outcome of immunotherapeutic strategies in cancer patients can be dependent on the gut microbiome (2). Treatment of cancer with check point inhibitors capable to unleash T cells to expand exponentially and kill transformed cells, is the newest and best treatment strategy being developed in the last decade. A true revolution in oncology is ongoing and we are seeing dramatic responses and long term overall survival in critical deadly tumors like advanced melanoma, non-small cell lung cancers (NSCLCs), renal cell carcinomas (RCCs). Not all patients respond and some of them show severe autoimmune toxicities. That response to immune checkpoint inhibitors (ICI) therapy, particularly anti-CTLA-4, could be associated to microbiome composition was first suggested by the group of Zitvogel in 2015 in melanoma (2). Other groups have confirmed and expanded the knowledges also by means of experimental models (3). Very recently the observation is being extended to epithelial cancers (4). Using shotgun DNA analysis performed on fecal samples from 60 NSCLC and 40 RCC patients before, during and after ICI therapy, they found an overrepresentation of the Firmicutes Akkermansia muciniphila present already at diagnosis in patients that with the therapy later showed a favorable clinical outcome. Moreover, antibiotics could compromise the efficacy of PD-1 blockade. Findings were further confirmed using the “avatar mice” model where mice were recolonized by fecal microbiota transplantation from responder and non-responder ICI treated patients. Restored immunity and antitumor activity of ICI could be achieved only with transplantation of stool samples from responder patients. Immunological mechanisms underlying these findings are still in the process of being fully elucidated. A specific Th1 response against A. muciniphila was detected associated with prolonged progression-free survival (PFS) in patients and in the antibiotic induced dysbiotic mice models the addition of Akkermansia was capable to increase T cell recirculation in lymph nodes and tumor beds. The close link with antitumor Th1 mediated immunity was already demonstrated since microbiota stimulated dendritic cells (DCs) are more efficient in activating T cell responses (5).
In other reports in the same issue of Science (6,7) melanoma patients microbiome was studied and a significant association was observed between commensal microbiome composition and clinical response of PD-1 treated patients. In the other report the amount of diversity of the microbiome repertoire appeared to be significant in defining ICI responder patients (Table 1).

Full table
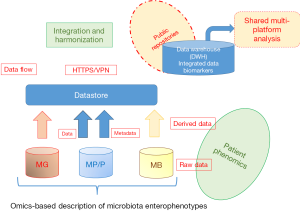
Significance of these findings is quite relevant for future strategies in oncology. First of all, the possibility to address dysbiotic indexes of microbiota maps, transferring translational research data into microbiome analytics, a new discipline, intended as microbiome “laboratory medicine”, serving as diagnostic tool for microbiome clinics. The next step of microbiota profiling need to exploit multi-omics approaches, also including metabolomics and proteomics in diagnostic algorithms to provide compositional but also functional maps of microbiota. From this diagnostic level, based on a new system medicine-based strategy, the possibility to manipulate microbiome with nutritional strategies, probiotics/prebiotics administration, eubiotic rather than antibiotic control or fecal transplantation can now be exploited to increase the prevalence of bacteria linked to anticancer immunosurveillance and to adapt bacteria ecosystem accordingly. It is necessary to build up reference biobanks of disease-associated microbiota profiles and fecal transplant dedicated material reservoirs. Variants of microbiota “enterophenotypes”, associated to treatments, clinical conditions and cancer types should be catalogued and stored into reference databases, both available for microbiomics scientists and for clinicians (Figure 1). Moreover, findings point out once more the important link with the development and manipulation of antitumor immunity. The immunotherapeutic strategies involving ICI that are nearly ready to go into the clinic will have to deal with these issues for the need to increase number of responder patients, identify optimal timing and schedule within standard therapies, decide as to whether perform microbiome testing for all cancer patients.
Acknowledgments
Funding: This work was supported by Associazione Italiana per la Ricerca sul Cancro (to M Nuti: AIRC IG 2015 Id.17432), Ricerche Universitarie (to M Nuti: Sapienza University 2017; to IG Zizzari: Sapienza University “Start up” 2017).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.25). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dinan TG, Cryan JF. Microbes, Immunity, and Behavior: Psychoneuroimmunology Meets the Microbiome. Neuropsychopharmacology 2017;42:178-92. [Crossref] [PubMed]
- Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079-84. [Crossref] [PubMed]
- Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084-9. [Crossref] [PubMed]
- Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91-7. [Crossref] [PubMed]
- Rescigno M A. 'fit' microbiota to potentiate cancer immunotherapy. Genome Med 2015;7:131. [Crossref] [PubMed]
- Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103. [Crossref] [PubMed]
- Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104-8. [Crossref] [PubMed]
- Putignani L, Del Chierico F, Vernocchi P, et al. Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood-Adulthood Transition. Inflamm Bowel Dis 2016;22:487-504. [Crossref] [PubMed]


