
Twist1 activated circRNA-10720 is a new player in hepatocellular carcinoma metastasis
Epithelial-mesenchymal transition (EMT) is a process that allows an epithelial cell to acquire a mesenchymal-like phenotype that enhances migration capacities. One of the first steps in the EMT process is the loss of cell-cell contacts, mediated in part by the loss of E-cadherin and other epithelial markers. Moreover, the acquisition of mesenchymal markers, such as Vimentin, is crucial in this process (1). EMT was first described in embryogenesis, where it plays a role in the formation of various tissues and organs, and later was related to oncology and the metastatic process (2). Twist1, a transcription factor first studied in embryonic development, where it acts as a morphogen regulating mesodermal differentiation (3), was first identified as an EMT gene by Yang et al. in 2004 (4). Twist1 acts as a transcriptional repressor of E-cadherin and has also been found to induce the expression of mesenchymal markers, such as Vimentin, Fibronectin and N-cadherin, during EMT (5). The role of Twist1 on EMT through upregulation of Vimentin levels in hepatocellular carcinoma (HCC) has been known since 2009 (6). However, the exact mechanism associated with the upregulation of Vimentin has not been elucidated, although transcriptional regulation has been ruled out since no binding site for Twist1 on the Vimentin promoter has been identified.
The recent study by Meng et al. (7) deciphers one of the mechanisms involved in the upregulation of Vimentin during the EMT process that enhances metastasis in HCC. The authors demonstrated that Twist1 activates the transcription of a CUL2-derived circular RNA (circRNA) that is involved in the absorption of the microRNAs (miRNAs) targeting Vimentin mRNA, hence producing a rise in Vimentin protein levels (Figure 1). CircRNAs are covalently closed single-strand RNA transcripts produced from a pre-mRNA mainly by two different mechanisms, back-splicing and exon-skipping (8). Their functions are mostly unknown, but some of them have been reported to regulate gene transcription and mRNA splicing and to act as a sponge for miRNAs affecting mRNA translation. In HCC, most of the circRNAs described to date have been shown to participate in the titration of miRNAs to regulate cell proliferation, apoptosis, cell cycle, different signaling pathways, invasion and metastasis (summarized in Table 1). However, the work of Meng et al. is the first that demonstrates in vitro and in vivo that a circRNA can regulate EMT in HCC through regulation of key molecules of the process such as Vimentin.
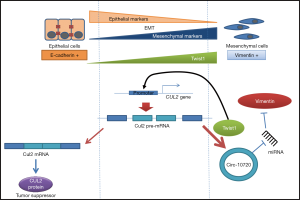
Table 1
| circRNA (ref) | circBase ID | Position (strand) | Host gene | HCC level | miRNA sponge in HCC | Function |
|---|---|---|---|---|---|---|
| circ-10720 (7) | hsa_circ_0018189 | chr10:35321362-35338693 (−) | CUL2 | Up | miR-1246, miR-127-5p, miR-331-5p, miR-1200, miR-888, miR-587, miR-656, miR-890, miR-490-5p, miR-1238, miR-548g, miR-513a-3p, miR-521 | Proliferation; Metastasis; EMT |
| ciRS-7/CDR1as (9) | hsa_circ_0001946 | chrX:139865339-139866824 (+) | CDR1 | Up | hsa-miR-7 | Proliferation; Metastasis |
| circHIPK3 (10,11) | hsa_circ_0000284 | chr11:33307958-33309057 (+) | HIPK3 | Up | miR-124, miR-152, miR-193a, miR-29a, miR-29b, miR-338, miR-379, miR-584, miR-654 | Proliferation; Metastasis |
| hsa_circ_0000673 (12) | hsa_circ_0000673 | chr16:11940357-11940700 (−) | RSL1D1 | Up | miR-767-3p | Proliferation; Metastasis |
| hsa_circ_0067934 (13) | hsa_circ_0067934 | chr3:170013698-170015181 (+) | PRKC1 | Up | miR-1324 | Proliferation; Metastasis |
| hsa_circ_0016788 (14) | hsa_circ_0016788 | chr1:228581376-228594517 (−) | TRIM11 | Up | miR-486 | Proliferation; Apoptosis; Metastasis |
| circRBM23 (15) | hsa_circ_0004137 | chr14:23375403-23378804 (−) | RBM23 | Up | miR-138 | Proliferation; Metastasis |
| hsa_circ_100338 (16) | – | chr1:151638888-151639119 (+) | SNX27 | Up | miR-141 | Metastasis |
| hsa_circ_000839 (17) | hsa_circ_0000497 | chr13:78293666-78327493 (+) | SLAIN1 | Up | – | Metastasis |
| hsa_circ_SLAIN1 (18) | hsa_circ_0100929 | chr13:78293666-78335245 (+) | SLAIN1 | Up | miR-375 | Proliferation; Apoptosis |
| hsa_circ_0005075 (19) | hsa_circ_0005075 | chr1:21377358-21415706 (−) | EIF4G3 | Up | miR-23b-5p, miR-93-3p, miR-581, miR-23a-5p | Proliferation; Metastasis |
| hsa_circ_0103809 (20) | hsa_circ_0103809 | chr15:51242247-51250991 (+) | AP4E1 | Up | miR-490-5p | Proliferation; Apoptosis; Metastasis |
| circFBLIM1 (21) | hsa_circ_0010090 | chr1:16084668-16113084 (+) | FBLIM1 | Up | miR-346 | Proliferation; Apoptosis; Metastasis |
| circ-ZEB1.33 (22) | hsa_circ_0004907 | chr10:31749965-31791437 (+) | ZEB1 | Up | miR-200a-3p | Proliferation |
| circRNA-101368 (23) | hsa_circ_0003028 | chr14:66028054-66028484 (+) | FUT8 | Up | miR-200a | Metastasis |
| hsa_circ_0078710 (24) | hsa_circ_0078710 | chr6:169625239-169654137 (−) | THBS2 | Up | miR-31 | Proliferation; Metastasis |
| circRNA_104075 (25) | hsa_circ_0075736 | chr6:17669523-17669777 (−) | NUP153 | Up | miR-582-3p | Proliferation |
| circSMAD2 (26) | hsa_circ_0000847 | chr18:45391429-45423180 (−) | SMAD2 | Down | miR-629 | Metastasis; EMT |
| circC3P1 (27) | – | chr19:10073090-10074135 (+) | C3P1 | Down | miR-4641 | Proliferation; Metastasis |
| hsa_circ_0005986 (28) | hsa_circ_0005986 | chr1:14057494-14068652 (+) | PRDM2 | Down | miR-129 | Proliferation |
| cSMARCA5 (29) | hsa_circ_0001445 | chr4:144464661-144465125 (+) | SMARCA5 | Down | miR-17, miR-181b | Proliferation; Apoptosis; Metastasis |
| circMTO1 (30) | hsa_circ_0007874 | chr6:74175931-74176329 (+) | MTO1 | Down | miR-9 | Proliferation; Apoptosis; Metastasis |
| circARSP91 (31) | hsa_circ_0085154 | chr8:101721360-101721451 (−) | PABPC1 | Down | – | Proliferation; Metastasis |
| circCDK13 (32) | hsa_circ_0001699 | chr7:40027197-40041630 (+) | CDK13 | Down | – | Metastasis |
| circZKSCAN1 (33) | hsa_circ_0001727 | chr7:99621041-99621930 (+) | ZKSCAN1 | Down | – | Proliferation; Metastasis |
| hsa_circ_0001649 (34) | hsa_circ_0001649 | chr6:146209155-146216113 (−) | SHPRH | Down | – | Proliferation; Apoptosis; Metastasis |
| circADAMTS14 (35) | hsa_circ_001866 | chr7:38295937-38305279 (−) | TARP | Down | miR-572 | Proliferation; Apoptosis; Metastasis |
To explore how Twist1 promotes EMT and specifically Vimentin upregulation, the authors performed chip-seq to identify Twist1 transcriptionally regulated genes and detected a binding site in the promoter of CUL2. CUL2 is a tumor suppressor gene involved in the ubiquitination and degradation of HIFα during normal vasculogenesis (36). When they analyzed the CUL2 levels in patient samples, no differences were observed between metastatic and non-metastatic patients, indicating that Twist1 action over CUL2 to promote EMT was not likely related to the coding form of the gene. Using a prediction model, seven potential circRNAs were identified that could be generated by back-splicing of CUL2 pre-mRNA, but only one—circ-10720—showed significant differences in expression between metastatic and non-metastatic HCC patients. Circ-10720 was significantly overexpressed in metastatic patients and its expression correlated with Twist1. Moreover, its expression was associated with disease stage and pathological grade, where advanced stages/grades showed higher levels, and also higher levels were found in patients with high fetoprotein (AFP) levels and with the hepatitis B marker. The expression of this CUL2-derived circRNA was clearly associated with a more aggressive phenotype in HCC patients and the patients showing high levels of circ-10720 had shorter overall survival.
The authors verified in vitro that Twist1 overexpression produces an increase of CUL2 pre-mRNA and circ-10720 levels, while CUL2 mRNA and protein levels were significantly reduced. The study of the potential oncogenic role of circ-10720 showed that after its in vitro upregulation, morphological changes could be observed in the cells with the acquisition of a mesenchymal-like phenotype and with upregulation of Vimentin and downregulation of E-cadherin levels, while the silencing of circ-10720 produced the reverse effect. Moreover, the upregulation of circ-10720 was associated with increased proliferation, migration and invasion. To study, how the CUL2-derived circRNA was regulating EMT, the authors then analyzed the potential role of this circRNA in miRNA titration. They showed that circ-10720 functions as a miRNA sponge and identified 14 miRNAs that bound to the circRNA, three of which (miR-1246, miR-578, miR-490-5p) had Vimentin as a target and were expressed in HCC, according to TCGA data. The upregulation of circ-10720 inhibits the miRNA repressive action over Vimentin mRNA translation, allowing upregulation of Vimentin protein levels, which are crucial for EMT. Moreover, the upregulation of circ-10720 produced higher molecular changes in the HCC cells, with an increase in cancer-associated and VEGF-associated genes in addition to the upregulation of EMT-associated genes and the downregulation of cell adhesion-related genes.
The authors performed several loss-of-function experiments to verify the Twist1—circ-10720 relation. In HCC cells overexpressing Twist1, which produces upregulation of Vimentin, the silencing of circ-10720 was associated with downregulation of Vimentin levels, which impacted proliferation, migration and invasion.
Using patient-derived tumor xenografts (PDTX) models, the authors monitored the growth of the primary tumors from different patients after in vivo modification of circ-10720 levels. The tumors were classified according to low or high Twist1 expression. In the group with low Twist1 levels, overexpression of circ-10720 through lentiviral transfection was associated with increased tumor volume. Inversely, in the group with high Twist1 levels, the silencing of circ-10720 was linked to reduced tumor growth. In both cases, the circRNA levels were correlated with the Vimentin levels.
Twist1 had previously been shown to participates in the EMT process, specifically by enhancing the intravasation step of metastasis, and the loss of Twist1 expression was directly related to a reduction of the number of circulating cells and a decrease in metastasis (4). Meng et al. (7) used a TetOn-Twist1 mouse model to evaluate the role of circ-10720 in metastasis. After activation of Twist1 overexpression, the mice first produced HCC tumors and later, after a long period of Twist1 activation, distant metastases emerged. Intravenous treatment with a siRNA against circ-10720 produced a reduction in metastasis. This model demonstrated the role of circ-10720 in metastasis, explained why the metastatic HCC patients had higher levels of circ-10720, and suggested a potential therapeutic use of this circRNA for the treatment of HCC.
Finally, the authors showed that circ-10720 can also be used as a prognostic biomarker in HCC. Detection of circ-10720 by FISH, together with Vimentin positivity, was an indicator of a more aggressive phenotype, higher risk of metastasis, and shorter overall survival. However, these results were obtained in a small cohort of 75 HCC patients and further investigation in an independent and larger cohort of patients is warranted to validate these findings.
In summary, this study is a good example of how circRNA expression can be regulated by cell type-specific mechanisms, as occurs during EMT when several circRNAs are activated (37). In the present work, CUL2 transcriptional processing was shown to be cell type-specific. In epithelial cells, CUL2 transcription produced an mRNA that will be translated to Cul2 protein, which acts as a tumor suppressor, while in mesenchymal cells, the overexpression of Twist1 promoted the transcription of a CUL2-derived circRNA produced by back-splicing through direct binding of Twist1 to the promoter region of CUL2. The CUL2-derived circRNA regulated the translation of Vimentin, thus inhibiting Vimentin-targeting miRNAs, and promoted tumor growth and metastasis (Figure 1). Further studies are needed to elucidate the other components that participate in the alternative processing of the CUL2 pre-mRNA that leads to CUL2-derived circRNA production during the EMT process, but the understanding of the Twist1-mediated activation of Vimentin is a significant step in the understanding of EMT and risk stratification in HCC and provides new potential therapeutic targets for HCC.
Acknowledgments
Funding:
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.12.01). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016. Cell 2016;166:21-45. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 1991;5:1568-76. [Crossref] [PubMed]
- Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004;117:927-39. [Crossref] [PubMed]
- Ansieau S, Morel AP, Hinkal G, et al. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene 2010;29:3173-84. [Crossref] [PubMed]
- Matsuo N, Shiraha H, Fujikawa T, et al. Twist expression promotes migration and invasion in hepatocellular carcinoma. BMC Cancer 2009;9:240. [Crossref] [PubMed]
- Meng J, Chen S, Han JX, et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res 2018;78:4150-62. [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Xu L, Zhang M, Zheng X, et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol 2017;143:17-27. [Crossref] [PubMed]
- Chen G, Shi Y, Liu M, et al. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 2018;9:175. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Jiang W, Wen D, Gong L, et al. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem Biophys Res Commun 2018;500:211-6. [Crossref] [PubMed]
- Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun 2018;497:626-32. [Crossref] [PubMed]
- Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol 2018;234:500-8. [Crossref] [PubMed]
- Wang B, Chen H, Zhang C, et al. Effects of hsa_circRBM23 on Hepatocellular Carcinoma Cell Viability and Migration as Produced by Regulating miR-138 Expression. Cancer Biother Radiopharm 2018;33:194-202. [Crossref] [PubMed]
- Huang XY, Huang ZL, Xu YH, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep 2017;7:5428. [Crossref] [PubMed]
- Wang BG, Li JS, Liu YF, et al. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol 2017;39:1010428317719577. [Crossref] [PubMed]
- Cao S, Wang G, Wang J, et al. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK 2. Immunol Cell Biol 2019;97:218-28. [Crossref] [PubMed]
- Shang X, Li G, Liu H, et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine (Baltimore) 2016;95:e3811. [Crossref] [PubMed]
- Cai H, Hu B, Ji L, et al. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res 2018;10:1690-702. [PubMed]
- Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res 2018;37:172. [Crossref] [PubMed]
- Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int 2018;18:116. [Crossref] [PubMed]
- Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle 2018;17:2349-59. [Crossref] [PubMed]
- Xie B, Zhao Z, Liu Q, et al. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene 2019;683:253-61. [Crossref] [PubMed]
- Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis 2018;9:1091. [Crossref] [PubMed]
- Zhang X, Luo P, Jing W, et al. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther 2018;11:2853-63. [Crossref] [PubMed]
- Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun 2018;499:1044-9. [Crossref] [PubMed]
- Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 2017;8:43878-88. [PubMed]
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 2018;68:1214-27. [Crossref] [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Ma Y, Zhang C, Zhang B, et al. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Available online: https://www.spandidos-publications.com/10.3892/ol.2018.9606
- Lin Q, Ling YB, Chen JW, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol 2018;53:246-56. [PubMed]
- Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol 2017;11:422-37. [Crossref] [PubMed]
- Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016;16:161-9. [Crossref] [PubMed]
- Song C, Li D, Liu H, et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J Cell Physiol 2019;234:2460-70. [Crossref] [PubMed]
- Maeda Y, Suzuki T, Pan X, et al. CUL2 is required for the activity of hypoxia-inducible factor and vasculogenesis. J Biol Chem 2008;283:16084-92. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]


