
A mechanism of regucalcin knock-down in the promotion of proliferation and movement of human cervical cancer HeLa cells
Introduction
The regucalcin (RGN) has been originally extracted from rat liver homogenate and named by Japanese scientists (1). It plays a key role in maintaining intracellular calcium homeostasis and liver metabolism. It is reported that RGN affects the activity of several enzymes such as pyruvate kinase, succinate kinase, glycogen phosphorylase and adenosine triphosphatase by the calcium ion (2-5). Recent studies showed that RGN can protect against radiation-induced testicular damage, regulate intracellular Ca2+ homeostasis in kidney proximal tubule epithelial cells and has a cryoprotective effect for buffalo spermatozoa in extender (6-8). Furthermore, more and more reports have demonstrated that the expression of RGN is decreased in cancer cells, such as liver cancer, breast cancer, prostate cancer, lung cancer, pancreatic cancer, colorectal cancer and renal cell carcinoma, compared to normal cells (9-18). Up-regulating RGN expression in the cancer cells above may inhibit proliferation, migration and other biological characteristics. In our previous study, siRNA has been transfected into HepG2 cells and cell proliferation and migration have shown to be promoted (9). In addition to liver cancer, cervical cancer is also one of the most common cancers in the world, especially in developing countries (19-21). The relationship between RGN and cervical cancer has not been reported. In this study, therefore, we have established two cell lines HeLa-siRGN and HeLa-NC by lentiviral siRNA and lentiviral NC. Cell proliferation, migration, invasion abilities and the expression of Wnt/β-catenin pathway and epithelial-mesenchymal transition (EMT) related proteins have been detected. The aim of this study is to provide theoretical basis for molecular targeted therapy of cervical cancer by targeting RGN.
Methods
Cell lines and lentivirus transfection
HeLa cells were supplied by the Chinese Academy of Sciences. HeLa cells were thawed in 37 °C water bath and cultured in Dulbecco’s modified eagle’s medium (DMEM) (Gibco, USA) with 10% fetal bovine serum (FBS) (Gibco, USA). The cells were seeded in a 6-well plate (5×105/well) and used for lentivirus infection. The multiplicity of infection (MOI) is 20. SiRNA for RGN interference was designed using Ambion company’s online design tool and then linked to lentivirus and packaged by GenePhama company, Suzhou, China. After 24 hours transfection, the virus diluent was removed and cells were incubated in puromycin solution (2 µg/mL) for 3 days to kill the cells failed in infection.
Quantitative real-time PCR
Total RNA was extracted by Total RNA extraction kit (Tiangen, China). PrimeScriptTM RT reagent Kit (TAKARA, Japan) was used for DNA removal and reverse transcription. Then, RGN mRNA expression was measured by SYBR Premix Ex TaqTM II (TAKARA, Japan). Primer sequences for RGN are as follows: forward, 5'-GTGGATGCCTTTGACTATGACC-3'; reverse, 5'-CTTCCCCTCAGCATCAATACAC-3'. Primer sequences for internal reference gene GAPDH are as follows: forward, 5'-CGAGATCCTCAACCAATCAA-3'; reverse, 5'-GGTGGTCCAGGGTCGTTACT-3'. All operations were carried out according to the kit instructions. The experimental results were processed by 2−ΔΔCt method.
Western blotting (WB)
Cells were collected with scrapers on ice-bath. The total protein was extracted with cell lysis buffer and protein concentration of each group was determined by bicinchoninic acid method (BCA, Beyotime, China). Forty µg proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. After blocking with skim milk, the membranes were incubated with polyclonal rabbit antibodies at 4 °C for a night and goat anti-rabbit IgG-HRP (1:5,000; E-AB-1003; Elabscience, China) at 37 °C for 1 hour. After washing the membranes with TBST for three times, the membranes were exposed and developed with Super ECL plus hypersensitive luminescence solution (Applygen, China). The relative expression of proteins was analyzed by Image J. The primary antibodies used are as follows: RGN (1:1,000; 17947-1-AP; Proteintech, USA), β-catenin (1:1,000; 51067-2-AP; Proteintech, USA), p-GSK-3β (1:1,000; ab75814; Abcam, UK), GSK-3β (1:1,000; 22104-1-AP; Proteintech, USA), MMP-3 (1:1,000; 17873-1-AP; Proteintech, USA), MMP-7 (1:800; 10374-2-AP; Proteintech, USA), MMP-9 (1:800; 10375-2-AP; Proteintech, USA), E-cadherin (1:1,000; 20874-1-AP; Proteintech, USA), N-cadherin (1:1,000; 22018-1-AP; Proteintech, USA), Vimentin (1:1,000; 10366-1-AP; Proteintech, USA), GAPDH (1:1,000; 10494-1-AP; Proteintech, USA).
Cell proliferation assay
Cells were seeded in five 96-well plates at a concentration of 3,000/well. Cell proliferation was determined at 0, 24, 48, 72 and 96 hours, respectively. The culture medium was aspirated and 100 µL fresh medium with 10 µL CCK-8 buffer was pipette to each well. The plates were incubated for about 4 hours and the absorbance of formazan produced in the chemical reaction can be detected at 450 nm using microplate reader. The amount of formazan is proportional to the number of living cells.
Colony formation assay
Five hundred cells of each group were added to the 6-well plates. During the time of culturing, the growth of cells was observed and fresh culture medium was replaced in time. After 14 days, the cells were rinsed twice with PBS, fixed with paraformaldehyde for 15 min and stained with 0.1% crystal violet for 10 min. Crystal violet was washed off with ddH2O and let the plates naturally dried. The number of colonies with cell number greater than 50 in each group was then counted.
Cell migration assay
Complete medium was added into the 24-well plates. Cells were collected to make cell suspension using DMEM and seeded in the upper chamber (3×104/well). Then, the plates were incubated at 37 °C for 24 hours to let the cells pass through the pores on the membrane. The cells on the upper layer of the chamber were gently erased, and the cells on the lower layer of the membrane were fixed with 100% methanol and stained with 0.1% crystal violet. Migratory cells were taken pictures, counted in 5 random fields (×100) and taken average values.
Cell invasion assay
Lay the 1:8 diluted matrigel onto the upper chamber of polycarbonate film and kept for a night at 37 °C. Cells were collected to make cell suspension using DMEM and seeded in the upper chamber (3×104/well). Then, the plates were incubated at 37 °C for 24 hours to let the cells pass through the pores on the membrane. The cells on the upper layer of the chamber were gently erased, and the cells on the lower layer of the membrane were fixed with 100% methanol and stained with 0.1% crystal violet. Invasive cells were taken pictures, counted in 5 random fields (×100) and taken average values.
Statistical analysis
Spss17.0 statistical software was used to process the data. Measurement data were expressed by mean ± SD and statistical analyses were performed using t-test. P<0.05 was considered to have statistically significant.
Results
RGN expression was down-regulated by siRGN
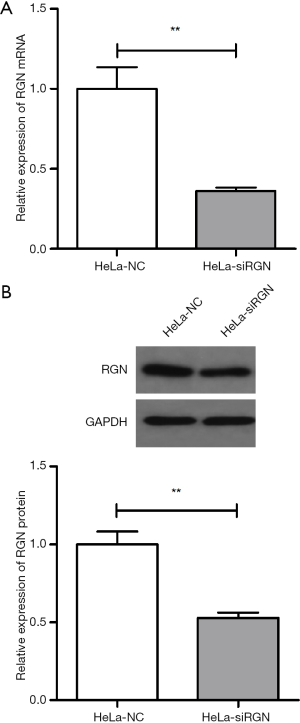
After lentivirus siRGN infection, the expression level of RGN mRNA and protein of HeLa-siRGN cell line were significantly lower than those of HeLa-NC cell line (P<0.01). The results demonstrated that the two cell lines HeLa-siRGN and HeLa-NC have been established successfully (Figure 1).
Down-regulation of RGN promoted proliferation
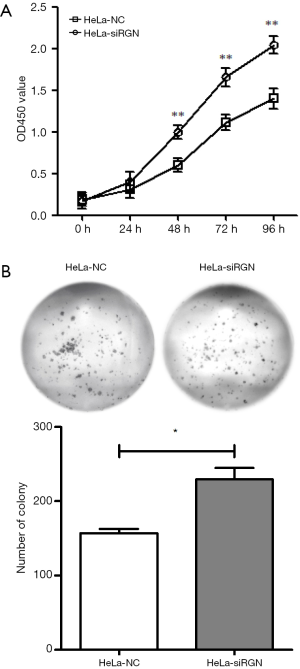
Figure 2A have shown that the proliferation rate of HeLa-siRGN cells was faster than that of HeLa-NC cells (P<0.01). From the results of clone formation experiment (Figure 2B), it can be seen that after 14 days of culture, the number of colonies (cell number greater than 50) in HeLa-siRGN group was more than what in HeLa-NC group (P<0.05).
Down-regulation of RGN enhanced cell migration and invasion
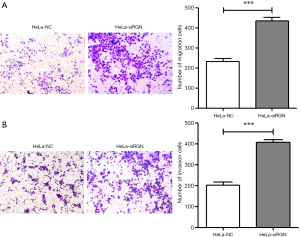
Cells were cultured in transwell chamber for 24 hours, and the cells passing through the membrane were observed by light microscope. Figure 3A shows that the average migratory cell number in HeLa-siRGN group was 434.33±28.54, in HeLa-NC group was 231.33±25.93 (P<0.001). Figure 3B shows that the average invasive cell number in HeLa-siRGN group was 405.67±24.13, in HeLa-NC group was 202.00±27.51 (P<0.001).
Down-regulation of RGN promoted the activation of Wnt/β-catenin pathway and EMT
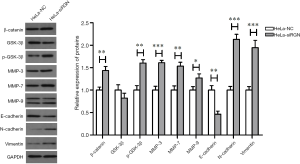
We have used WB to detect the effect of RGN down-regulation on Wnt/β-catenin pathway and EMT. Figure 4 shows that the expression of E-cadherin in HeLa-siRGN decreased (P<0.01) and the expression of β-catenin, p-GSK-3β, MMP-3, MMP-7, MMP-9, N-cadherin and vimentin increased (PMMP-9<0.05, Pβ-catenin, p-GSK-3β, MMP-7<0.01, PMMP-3, N-cadherin, vimentin<0.001).
Discussion
The role of RGN in tumor has been paid more and more attention by researchers. Its anti-tumor effect may make it a potential target for cancer gene therapy. This study has shown that RGN down-regulation can significantly promote the proliferation, migration and invasion abilities of HeLa cells. Furthermore, RGN down-regulation has decreased the expression of E-cadherin and increased the expression of β-catenin, p-GSK-3β, MMP-3, MMP-7, MMP-9, N-cadherin and vimentin. From the above results, it seems that, after down-regulation of RGN, the Wnt/β-catenin pathway of HeLa cells has been activated, and the EMT process has been promoted.
In 1990s, several researchers have found that exogenous double-stranded RNA injected into nematodes is able to induce homologous gene silencing and it has been called RNA interference (22). The SiRNA has the abilities to specifically recognize homologous genes and selectively inhibit gene expression when transferred into cells by proper vector. It makes siRNA a good way in studying specific genes (23,24). Nowadays, many developing countries still have a high incidence of cervical cancer due to inadequate screening and prevention. Gene therapy is undoubtedly one of the most promising methods in cervical cancer treatment. A common method to know a gene is to down-regulate it by siRNA. Many researchers have demonstrated that RGN can weaken the growth and migration of cancer cells. It is reported that overexpressing RGN suppresses the proliferation of rat hepatoma H4-II-E cells, which was approached by suppressing various protein kinase activities such as Ca2+/calmodulin-dependent protein kinase, protein kinase C, and protein tyrosine kinase (10-11,25). It can also suppress the growth of liver cancer cells HepG2 when culturing it with medium supplemented with regucalcin (12). Ricardo Marques established transgenic rats overexpressing RGN (Tg-RGN) and found Tg-RGN rats had lower incidence of carcinogen-induced mammary gland tumors, lower proliferation of tumor cells, and less invasive forms, compared to wild type rats (13). Furthermore, Masayoshi Yamaguchi has found overexpression of RGN can suppressing cell proliferation, death and migration when transfected RGN cDNA into non-small-cell lung cancer (NSCLC) A549 cells, human colorectal carcinoma RKO cells and human clear cell RCC A498 cells (15-17). Our study is to investigate if the biological characteristics can reverse when endogenous RGN expression is down-regulated, compared to RGN transfection cancer cells. As expected, our study shows that the proliferation, migration and invasion abilities of HeLa-siRGN cells have increased significantly. It has been demonstrated that the proliferation suppression effect of RGN is due to the inhibition of various signaling pathways such as Akt, MAPK, SAPK/JNK and Ras (15,16,18,26). In our research, RGN under-expression enhanced cell migration significantly. So, we have studied the effect of RGN on the signaling pathways proteins associated with metastasis. E-cadherin, vimentin and N-cadherin are markers of EMT (27-29). Our studies have shown that the expression of E-cadherin in HeLa-siRGN cells decrease and the expression of vimentin and N-cadherin increase, indicating that the EMT process has been activated. Furthermore, by detecting the key genes of the Wnt/β-catenin pathway, it has been found that RGN down-regulation led to the increase of p-GSK-3β expression. This means that GSK-3β activity has reduced, and less β-catenin has been degraded by ubiquitination, which leads to excessive β-catenin entering the nucleus, forming more β-catenin/LEF/TCF complex, promoting the transcription and expression of downstream genes such as MMP3\MMP7\MMP9, and promoting the invasion and migration of HeLa cells (30-35). It seems that deficiency of RGN can promote cervical cancer. Reports have shown that RGN mRNA and protein expression was diminished in breast and prostate cancer tissues, and RGN expression was negatively correlated with the histological grade of breast infiltrating ductal carcinoma (13,14). The studies above seem to suggest that the loss of RGN can make various cancers to get worse. While, higher RGN expression has been seen in lung cancer and pancreatic ductal adenocarcinomas patients with prolonged survival (15,18). In addition, RGN overexpression decreases the oncogenes c-fos, c-myc, Ha-ras and elevation of the tumor suppressers p53 and Rb (15,36). It is no doubt that RGN is a protective factor in cancer progression (37).
In conclusion, our study shows the decreased expression of RGN promotes the progression of cervical cancer and its effect is probably achieved by activating Wnt/β-catenin pathway and EMT. Our research provides the theoretical basis of RGN for anti-cancer molecular therapy.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.02.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional Review Board review was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yamaguchi M, Sugii K. Properties of calcium-binding protein isolated from the soluble fraction of normal rat liver. Chem Pharm Bull (Tokyo) 1981;29:567-70. [Crossref] [PubMed]
- Yamaguchi M, Shibano H. Effect of calcium-binding protein on the activation of phosphorylase a in rat hepatic particulate glycogen by Ca2+. Chem Pharm Bull (Tokyo) 1987;35:2581-4. [Crossref] [PubMed]
- Yamaguchi M, Shibano H. Calcium-binding protein isolated from rat liver cytosol reverses activation of pyruvate kinase by Ca2+. Chem Pharm Bull (Tokyo) 1987;35:2025-9. [Crossref] [PubMed]
- Yamaguchi M, Shibano H. Reversible effect of calcium-binding protein on the Ca2+-induced activation of succinate dehydrogenase in rat liver mitochondria. Chem Pharm Bull (Tokyo) 1987;35:3766-00. [Crossref] [PubMed]
- Yamaguchi M, Mori S, Kato S. Calcium-binding protein regucalcin is an activator of (Ca2+-Mg2+)-adenosine triphosphatase in the plasma membranes of rat liver. Chem Pharm Bull (Tokyo) 1988;36:3532-39. [Crossref] [PubMed]
- Silva AM, Correia S, Casalta-Lopes JE, et al. The protective effect of regucalcin against radiation-induced damage in testicular cells. Life Sci 2016;164:31-41. [Crossref] [PubMed]
- Yamaguchi M. The potential role of regucalcin in kidney cell regulation: Involvement in renal failure Int J Mol Med 2015;36:1191-9. (Review). [Crossref] [PubMed]
- Pillai H, Parmar MS, Shende AM, et al. Effect of supplementation of recombinant Regucalcin in extender on cryopreservation of spermatozoa of water buffalo (Bubalus bubalis). Mol Reprod Dev 2017;84:1133-9. [Crossref] [PubMed]
- Zhang SC, Liang MK, Huang GL, et al. Inhibition of SMP30 gene expression influences the biological characteristics of human Hep G2 cells. Asian Pac J Cancer Prev 2014;15:1193-6. [Crossref] [PubMed]
- Inagaki S, Yamaguchi M. Suppressive role of endogenous regucalcin in the enhancement of protein kinase activity with proliferation of cloned rat hepatoma cells (H4-II-E). J Cell Biochem Suppl 2001;12-8. [Crossref] [PubMed]
- Inagaki S, Yamaguchi M. Regulatory role of endogenous regucalcin in the enhancement of nuclear deoxyribonuleic acid synthesis with proliferation of cloned rat hepatoma cells (H4-II-E). J Cell Biochem 2001;82:704-11. [Crossref] [PubMed]
- Yamaguchi M, Murata T. Exogenous regucalcin suppresses the growth of human liver cancer HepG2 cells in vitro. Oncol Rep 2018;39:2924-30. [PubMed]
- Marques R, Vaz CV, Maia CJ, et al. Histopathological and in vivo evidence of regucalcin as a protective molecule in mammary gland carcinogenesis. Exp Cell Res 2015;330:325-35. [Crossref] [PubMed]
- Maia C, Santos C, Schmitt F, et al. Regucalcin is under-expressed in human breast and prostate cancers: Effect of sex steroid hormones. J Cell Biochem 2009;107:667-76. [Crossref] [PubMed]
- Yamaguchi M, Osuka S, Shoji M, et al. Survival of lung cancer patients is prolonged with higher regucalcin gene expression: suppressed proliferation of lung adenocarcinoma A549 cells in vitro. Mol Cell Biochem 2017;430:37-46. [Crossref] [PubMed]
- Yamaguchi M, Osuka S, Murata T. Prolonged survival of patients with colorectal cancer is associated with a higher regucalcin gene expression: Overexpression of regucalcin suppresses the growth of human colorectal carcinoma cells in vitro. Int J Oncol 2018;53:1313-22. [PubMed]
- Yamaguchi M, Osuka S, Hankinson O, et al. Prolonged survival of renal cancer patients is concomitant with a higher regucalcin gene expression in tumor tissues: Overexpression of regucalcin suppresses the growth of human renal cell carcinoma cells in vitro. Int J Oncol 2019;54:188-98. [PubMed]
- Yamaguchi M, Murata T. Suppressive effects of exogenous regucalcin on the proliferation of human pancreatic cancer MIA PaCa-2 cells in vitro. Int J Mol Med 2015;35:1773-8. [Crossref] [PubMed]
- Markman M. Advances in cervical cancer pharmacotherapies. Expert Rev Clin Pharmacol 2014;7:219-23. [Crossref] [PubMed]
- Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets 2013;13:205-20. [Crossref] [PubMed]
- Marth C, Landoni F, Mahner S, et al. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv262. [Crossref] [PubMed]
- Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806-11. [Crossref] [PubMed]
- Mello CC, Conte D. Revealing the world of RNA interference. Nature 2004;431:338-42. [Crossref] [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Misawa H, Inagaki S, Yamaguchi M. Suppression of cell proliferation and deoxyribonucleic acid synthesis in the cloned rat hepatoma H4-II-E cells overexpressing regucalcin. J Cell Biochem 2001;84:143-9. [Crossref] [PubMed]
- Yamaguchi M, Osuka S, Weitzmann MN, et al. Prolonged survival in pancreatic cancer patients with increased regucalcin gene expression: Overexpression of regucalcin suppresses the proliferation in human pancreatic cancer MIA PaCa-2 cells in vitro. Int J Oncol 2016;48:1955-64. [Crossref] [PubMed]
- Cong N, Du P, Zhang A, et al. Downregulated microRNA-200a promotes EMT and tumor growth through the wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol Rep 2013;29:1579-87. [Crossref] [PubMed]
- Xu HG, Zheng Q, Song JX, et al. Intermittent cyclic mechanical tension promotes endplate cartilage degeneration via canonical Wnt signaling pathway and E-cadherin/β-catenin complex cross-talk. Osteoarthritis Cartilage 2016;24:158-68. [Crossref] [PubMed]
- Zhang X, Yang M, Shi H, et al. Reduced E-cadherin facilitates renal cell carcinoma progression by WNT/β-catenin signaling activation. Oncotarget 2017;8:19566-76. [PubMed]
- Dong ZC, Zhang D, Wang SB, et al. Target inhibition on GSK-3β by miR-9 to modulate proliferation and apoptosis of bladder cancer cells. Eur Rev Med Pharmacol Sci 2018;22:3018-26. [PubMed]
- Song B, Lin HX, Dong LL, et al. MicroRNA-338 inhibits proliferation, migration, and invasion of gastric cancer cells by the Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2018;22:1290-6. [PubMed]
- Kypta RM, Waxman J. Wnt/β-catenin signalling in prostate cancer. Nat Rev Urol 2012;9:418-28. [Crossref] [PubMed]
- Burgy O, Königshoff M. The WNT signaling pathways in wound healing and fibrosis. Matrix Biol 2018;68-69:67-80. [Crossref] [PubMed]
- Bahrami A, Hasanzadeh M. Clinical Significance and Prognosis Value of Wnt Signaling Pathway in Cervical Cancer. J Cell Biochem 2017;118:3028-33. [Crossref] [PubMed]
- Jacques BE, Montgomery WH, Uribe PM, et al. The role of Wnt/β-catenin signaling in proliferation and regeneration of the developing basilar papilla and lateral line. Dev Neurobiol 2014;74:438-56. [Crossref] [PubMed]
- Tsurusaki Y, Yamaguchi M. Overexpression of regucalcin modulates tumor-related gene expression in cloned rat hepatoma H4-II-E cells. J Cell Biochem 2003;90:619-26. [Crossref] [PubMed]
- Vaz CV, Correia S, Cardoso HJ, et al. The Emerging Role of Regucalcin as a Tumor Suppressor: Facts and Views. Curr Mol Med 2016;16:607-19. [Crossref] [PubMed]


