
The molecular basis for p53 inhibition of autophagy in porcine fibroblast cells
Introduction
Autophagy is an evolutionarily conserved homeostatic process that degrades and recycles cytosolic components in autophagosomes (1). When cells are subjected to various stress signals, including nutrient deprivation, autophagy is usually activated (2). This dynamic process is highly controlled by autophagy-related (ATG) genes which encode autophagosome formation (3). The core process of autophagy is categorized into five steps: (I) autophagosome formation initiated by the ATG1/ULK1 complex (4); (II) nucleation regulated by the ATG12 conjugation system (5); (III&IV) elongation regulated by the ATG8/LC3 conjugation/deconjugation system and beclin-1/PI3K complex (6); and (V) degradation by the ATG9/ATG9L1 cycling system (7). Additionally, autophagy also regulated by signaling pathways, including p53, PI3K, RAS and JAK-STAT signaling pathways (8). Therefore, autophagy regulation is an intricate network, and the molecular basis for it has not been fully elucidated.
p53, a well-known tumor suppressor, plays an important role in cellular genome stability (9). In response to a myriad of stresses, such as starvation stress, genotoxic stress and other forms of stress, p53 can induce diverse cellular responses, including cell cycle arrest, apoptosis and autophagy (10). Numerous studies have reported that p53 plays a dual role in the regulation of autophagy (11,12).
It has been reported that autophagy is regulated by p53 through signaling pathways in a transcription-dependent or transcription-independent manner. When p53 is activated, autophagy may be induced by inhibiting the mammalian target of rapamycin (mTOR) pathway and activating the AMP-responsive protein kinase (AMPK) pathway (13). Additionally, p53 may also induce autophagy by activating target genes that code for pro-autophagic modulators, including PTEN, DRAM, IGF-BP3, and ARF, and pro-apoptotic Bcl-2 proteins (Bax, PUMA) (10). Strikingly, p53 and TP53-induced glycolysis and apoptosis regulator (TIGAR) may inhibit autophagy directly (14). Several studies indicated that p53 can serve as either an inducer or inhibitor, depending on the cellular context, type of stress and the subcellular localization of p53 (15). However, the mechanism by which p53 regulates autophagy still need to be elucidated.
Previous studies have demonstrated that the deletion or inhibition of p53 induced an autophagy response in human, mouse and nematode cells (16). However, the relationship between autophagy and p53 in PFCs need to be identified. In this study, we investigated the role of p53 in the regulation of autophagy and the molecular difference in the autophagic response in starvation-induced autophagy in PFCs.
Methods
Reagents
Earle’s balanced salt solution (EBSS) was purchased from Gibco (St. Louis, MO, USA). Rabbit anti-p62 (Cat# P0067), rabbit anti-LC3B (Cat# L7543) and mouse anti-β-actin (Cat#A5541) were obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Bafilomycin A1 (Baf A1) was purchased from Sangon Biotech (Shanghai, China).
Cell lines and cell culture
Wild type p53 (p53wt) and p53 knockout (p53-/-) PFCs were established and the p53 expression status was confirmed as described in our previous study (17). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, NY, USA), 100 IU/mL penicillin and 100 IU/mL streptomycin. All cells were incubated in a humidified 5% CO2 atmosphere at 37 °C.
Protein extraction and immunoblotting
Protein extraction and immunoblotting were performed according to our previous study (18). Briefly, after treatment, the cells were washed twice with PBS and collected. Then, the cell lysate concentration was determined using a BCA protein assay kit (Beyotime, Shanghai, China). Next, 50 µg of the total protein samples was separated by 12% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% BSA (Solarbio, Beijing, China) for 2 h and then incubation with a diluted primary antibody (LC3B: 1:2,000, p62: 1:2,000, β-actin: 1:5,000) overnight at 4 °C. Subsequently, the membranes were washed and incubated with the appropriate secondary antibodies (R&D, 1:5,000, USA) for 2 h at room temperature. Finally, the membranes were incubated with ECL (Easysee Western Blotting Kit, Transgenes, China) and visualized with an imaging system (Bio-Rad, Hercules, USA).
Transmission electron microscopy (TEM)
Samples used for transmission electron microscopy (TEM) analysis were harvested and washed twice with cold PBS. Before dehydrated in ethanol, the samples were fixed in 2.5% glutaraldehyde for 30 min at room temperature and incubated overnight at 4 °C. The samples were washed three times with a 0.1 M phosphoric acid buffer solution and post-fixed with 1% osmium tetroxide for 2–3 h at 4 °C. Then, the samples were treated with a mixed solution of acetone and embedding solution and embedded in Spurr’s resin for the preparation of ultrathin sections. After staining with 3% uranyl acetate and lead citrate, ultrathin sections were examined using a transmission electron microscope (JEM 1011, JEOL, Japan).
Immunofluorescence
The p53wt and p53-/- PFCs (5×104 cells/well) were seeded on 6-chamber culture slides and pretreated with Baf A1 (100 nM) for 2 h, followed by treated with EBSS for 2 h. Then, the cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with PBS containing 5% BSA and 0.3% Triton X-100 for 30 min. After that, the cells were incubated with anti-LC3B antibody (1:200 diluted in 1.5% BSA) at 4 °C overnight. Cells were then incubated for 1 h with 1:400 secondary FITC-conjugated antibody, followed by incubation with DAPI for nuclear staining. Images were captured with a confocal fluorescence microscope (OLYMPUS FV 1000, Tokyo, Japan).
Acridine orange (AO) staining
To further detect the acidic vesicular organelles (AVOs), vital staining of PFCs with acridine orange (AO) (Sigma-Aldrich, St. Louis, MO, USA) was performed. Briefly, after treatment as described above, p53wt and p53-/- PFCs were stained with AO (5 µg/mL) for 15 min, washed with PBS and then visualized with fluorescence microscope (OLYMPUS IX71, Tokyo, Japan).
Quantitative real-time polymerase chain reactive (q-PCR) assay
p53wt and p53-/- PFCs were treated with EBSS for 2 h. Total RNA was extracted by Trizol (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from the total RNA using a Prime-Script RT reagent kit (TaKaRa, Tokyo, Japan). The obtained cDNA was used as a template for SYBR Green-based q-PCR (CFX-96, Bio-Rad, Hercules, CA, USA). The mRNA expression levels of the ATG genes were assessed with quantitative polymerase chain reaction (q-PCR). GAPDH was used for normalization. The primers used are shown in Table 1.
Table 1
| Gene | Primers sequence (5’ to 3’) | |
|---|---|---|
| Forward | Reverse | |
| ULK1 | F: CTCCCAGAGTGTCCCTCAAATGC | R: TGCTCCTCTTCTTCTACGTCCAAGC |
| ULK2 | F: AGAACAAGCAGAAGCTCCGAAAGC | R: AGAGCCAATGATTGTGGGTAAAGGA |
| VPS15 | F: TGGCAGCCTTGGGCATAACT | R: GAATCGCCTCCTGGTCACTCC |
| PIK3C3 | F: CAAGTGAGAATGGTCCGAATG | R: GTGGAAGAGTTTGCCTGTTTT |
| BECN1 | F: CAATAACTTCAGGCTGGGTCG | R: CGGCAGCTCCTTAGATTTGTCT |
| P62 | F: CTGTGGCTAGTGGTGGTTGGC | R: CTCGTGGTCGCTGAAGTCTGG |
| ATG4A | F: GAGCCAATCACCGTTTCTCAG | R: CGTCCACCTCCTCATTTCAGT |
| ATG4B | F: CACCGTCGCCCAGGTTCTCAA | R: GGTCGGAATCTGCGGGAAAGG |
| ATG10 | F: TTCTTCCCTTCTGCTCATCTT | R: CTGTCTTCGCTCAGTAGGTTAA |
| ATG16L1 | F: GAAGGAACTCGCAGAAGCAGC | R: CGCCTCCCAAAGATATTAGTGATAGA |
| ATG2A | F: TTCTTCCAGGAGCACCTCAGCC | R: CAGCCACCTCGATAGAGCCCAC |
| ATG2B | F: TGAGTGACGCTATGGAGGAGA | R: CAAGTGGTGAGTGAATGAGGC |
| ATG9A | F: CAAGAAGTACAGGCCCGGATTGTG | R: CGCAGAGGCAGGAGGGATTTG |
| ATG9B | F: GCGGCTTTGCCTGTATCCTGC | R: CATCCGACAAGGTCACTTTGCTGTG |
| mTOR | F: GTCTGCTGATGGGAGAATGGC | R: GTGAGGTAATGAGATGGGTGAAGG |
| EPG5 | F: TTCAGCATCTTCTTGGGTTCA | R: TTCGCTTGACAAACTGCCTAT |
| FOXO1 | F: ACCGCTTTACAAGTGCCTCTGC | R: GCTCAATGAACATGCCATCCAA |
| LMNA | F: CGAGGTCAAGTGGGTGTAGGAGG | R: GATGGGAAACACGAGGCAAGG |
| AMBRA1 | F: TGTTGCTCGCTCCTTCTTTTCCTGT | R: ACAGGAAAAGAAGGAGCGAGCAACA |
| DRAM1 | F: ATACAGGAACAACACCTCCAGAAAG | R: CCAACCCAAGAACTAGCGACA |
| GAPDH | F: ATCAAGAAGGTGGTGAAGCAG | R: CAGCATCAAAAGTGGAAGAGTG |
Measurement of autophagy flux
Autophagy flux was assessed in p53wt and p53-/- PFCs. A difference in LC3B-II levels in the presence and absence of lysosomal degradation (Baf A1, 100 nM) represents autophagic flux (19). Autophagy flux (AF) was calculated by the following equations according to the method of Gupta et al. (20): UT AF = (UT + Baf A1) − (UT − Baf A1), MT AF = (MT + Baf A1) − (MT − Baf A1), and ∆AF = (MT AF) − (UT AF), where UT is untreated and MT is modulator (EBSS)-treated.
Statistical analysis
Comparisons were performed using Student’s t-test. Quantitative data are expressed as the means ± SD. *P<0.05 and **P<0.01 versus the control were considered significant.
Results
Effect of starvation on autophagy in p53wt and p53-/- porcine fibroblasts cells
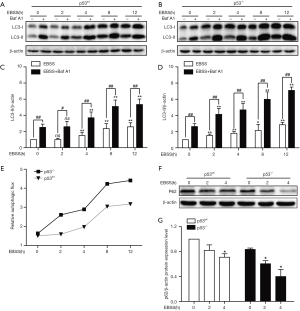
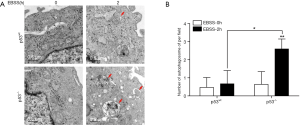
Increased expression of LC3-II and decreased expression of P62 are considered autophagosomal markers, but increases in LC3-II can be caused by increased autophagic flux or can be a result of blocking in autophagosome-lysosome fusion or degradation (19). To distinguish between these two possibilities, we performed an autophagy flux assay using the lysosomal ATPase inhibitor, Baf A1, which blocks lysosomal acidification. Immunoblotting results showed the expression of LC3B-II increased in a time-dependent manner in both groups of treated PFCs. This effect was further augmented by BafA1 treatment (Figure 1A,B,C,D). The digital results show that the expression of LC3B-II was significantly increased at 4 h in treated p53-wt PFCs, while the expression of LC3B-II was significantly increased at 2 h in p53-/- PFCs (Figure 1B,D). Moreover, the relative autophagic flux of p53-/- PFCs was stronger than that of p53wt PFCs at 2 h (Figure 1E). Meanwhile, we also found the expression of p62 decreased in a time-dependent manner in both groups of treated PFCs (Figure 1F,G). In addition, TEM results showed accumulation of autophagosomes production in p53-/- PFCs than in p53wt PFCs (Figure 2). These results demonstrated that starvation induced autophagy in both p53wt and p53-/- PFCs at different time points, and the initiation time of the autophagy response was earlier in p53-/- PFCs than in p53wt PFCs, which implies that p53 knockout induced autophagy without affecting lysosomal degradation in response to starvation.
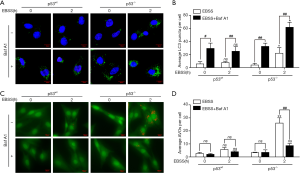
Effect of starvation on the formation of LC3 punctate and AVOs in p53wt and p53-/- porcine fibroblasts cells
AVOs can be stained by AO dye and emits bright red fluorescence, which are considered as the indicative of autophagy (21). To further ensure that starvation induced autophagy in p53wt and p53-/- PFCs, LC3 punctate, AVOs were monitored and quantified. As shown in Figure 3A, the number of LC3 punctate was significantly increased in the presence of EBSS in both group of PFCs. Moreover, the number of LC3 punctate was significantly increased after treatment with EBSS or combination with EBSS and Baf A1for 2 h in p53-/- PFCs, while the number of LC3 punctate has no effect in p53wt PFCs. In addition, the formation of AVOs was significantly increased after treated with EBSS for 2 h in p53-/- cells, while the formation of AVOs was significantly decreased in the presence of Baf A1. Moreover, the formation of AVOs was no change after treatment with EBSS or combination with EBSS and Baf A1 for 2 h (Figure 3B). These results further confirmed that p53 knockout induced an increasing autophagy flux without affecting lysosomal degradation in response to starvation, while the autophagy flux of p53wt PFCs had no change.
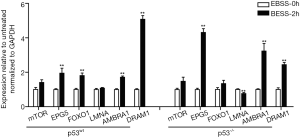
Effect of starvation on the mRNA expression level of autophagy signaling pathway-related genes in p53wt and p53-/- porcine fibroblasts cells
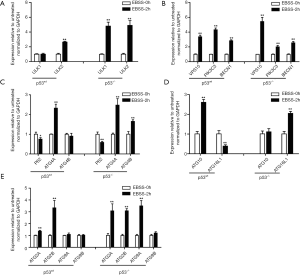
The genes upstream of autophagy, including FOXO1, mTOR, EPG5, LMNA, AMBRA1 and DRAM1, induce autophagy through signaling pathways (22-24). We found that the mRNA expression levels of the autophagy signaling pathway-related genes EPG5, AMBRA1 and DRAM1 were significantly increased in both types of cells; the expression of the LMNA gene was down-regulated significantly in p53-/- PFCs, but was unchanged in p53wt PFCs. In addition, the expression of the FOXO1 gene was up-regulated significantly in p53wt PFCs, but unchanged in p53-/- PFCs (Figure 4). These results demonstrated that the partial autophagy signaling pathway response might be different in the two types of PFCs.
Effect of starvation on the mRNA expression level of autophagy-related genes in p53wt and p53-/- porcine fibroblasts cells
Autophagy is a classic and highly conserved catabolic process, that includes initiation, nucleation, elongation and degradation cycling, which are controlled by autophagy-related genes. The initiation of autophagy begins with activation of the ULK1 complex, including ULK1, ULK2, ATG13, FIP200 (also known as RB1CC1) and ATG101, which activates nucleation-related genes. A class III PI3K complex (VPS15, PIK3C3 and BECN1), ATG4A, ATG4B, ATG10 and ATG16L1 are conducive to elongation, and ATG2A, ATG2B, ATG9A and ATG9B are the degradation cycling-related genes (25). To determine the autophagy-related gene response of both types of treated PFCs, we detected the mRNA expression levels of the autophagy-related genes described above by qPCR. We found the following: the mRNA expression levels of the ULK1, ULK2, VPS15, PIK3C3, BECN1, ATG4A, ATG4B, ATG16L1, ATG9A, ATG2A and ATG2B genes were significantly increased, and p62 was significantly decreased after treated EBSS at 2h than treated with EBSS at 0 h in p53-/- PFCs (Figure 5). The mRNA expression levels of the ULK2, VPS15, PIK3C3, BECN1, ATG4A, ATG2A, ATG2B and ATG10 genes were significantly increased at 2 h, whereas the mRNA expression levels of the ATG16L1 and p62 genes were significantly decreased after EBSS treated at 2 h than EBSS treated at 0h in p53wt PFCs (Figure 5). Interestingly, we found that the expression of the ULK1, ATG4B and ATG9A genes was up-regulated in p53-/- PFCs, while these genes had no change in p53wt PFCs. Additionally, the expression of ATG16L1 was up-regulated in p53-/- PFCs but down-regulated in p53wt PFCs. These results demonstrated that the autophagy-related gene response in the process of autophagosome formation was distinctly different in both types of treated PFCs.
Discussion
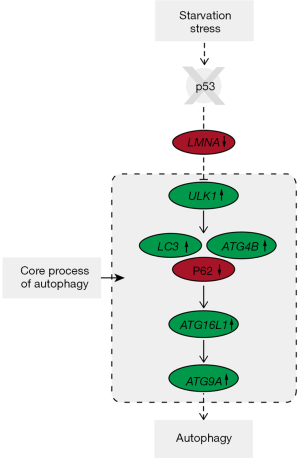
The relationship between p53 and autophagy regulation is an intricate one. Some previous studies have shown that p53 positively regulates autophagy (26), but downregulation of p53 induces autophagy in human trophoblast differentiation (27). Tasdemir et al. indicated that pharmacological inhibition and knockdown or knockout of p53 can induce autophagy in mouse and nematode cells (28). Our results were consistent with this finding. In this study, we found starvation-induced autophagy in p53wt and p53-/- PFCs (Figures 1-3), implying that p53 negatively regulates autophagy in response to starvation. Interestingly, we found that the initiation time of the autophagy response was earlier in p53-/- PFCs than in p53wt PFCs (Figure 1). It has been reported that autophagy is regulated by Ras signaling or JAK-STAT signaling under nutrient depletion (8). The p53-mediated autophagy may be related to cell type, stress-specific or activation of other p53 signaling pathway, which need further study.
The LMNA gene, which encodes lamin A and C (lamin A/C), could inhibit autophagy by activating mTOR in a mouse model (22). Of the autophagy signaling pathway-related genes, LMNA gene levels were lower in treated p53-/- PFCs than in treated p53wt PFCs, while FOXO1 gene levels were higher in treated p53wt PFCs than in treated p53-/- PFCs (Figure 4). These findings suggested that p53 may negatively regulate autophagy in a transcription-independent manner through targeting the LMNA/AKT/mTOR signaling pathway and positively regulate autophagy by targeting the PI3K/AKT/FOXO1 signaling pathway. Consistently, some studies also reported that the LMNA/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways may regulate autophagy in MCF-7 and MDA-MB-231 breast cancer cells and in other cells (18,29). Therefore, the role of p53 in controlling autophagy in PFCs requires further study.
The molecular basis of autophagy has been studied extensively in the setting of starvation stress, in which ATG proteins induction of autophagosome formation is considered basic research. ULK1 is a serine/threonine kinase that plays a role in autophagy by recruiting downstream ATG proteins (30). The ATG4B, ATG9 and ATG16L1 genes are essential for autophagosome elongation, maturation and degradation (31). In this study, we found that the levels of the ULK1, ATG4B and ATG9A genes were higher in treated p53-/- PFCs than in treated p53wt PFCs. In addition, the ATG16L1 gene was downregulated in treated p53wt PFCs but upregulated in treated p53-/- PFCs (Figure 5). It has been indicated that the dephosphorylation of ULK1 at Ser637 occurs in a p53-dependent manner in p53-null mouse embryonic fibroblasts (32). These findings suggest that p53 may negatively regulate autophagy in a transcription-independent manner through targeting the ULK1, ATG4B, ATG16L1 and ATG9A genes. However, the specific mechanism involved in targeting the ULK1, ATG4B, ATG16L1 and ATG9A genes requires further study.
Conclusions
In conclusion, our findings indicated that p53 negatively regulated autophagy under starvation conditions in PFCs, the autophagic flux of p53-/- PFCs increased without affecting lysosomal degradation in response to starvation in early phase. Furthermore, we hypothesized that targeting ATG genes and signaling pathways via p53 knockout would induce a response to starvation. A schematic representation of the proposed mechanism of autophagy regulation by p53 knockout in PFCs proposed in the present study is shown in Figure 6. These findings will provide a new biological model and support further study of the molecular mechanism of p53 in controlling autophagy in response to starvation.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.05.22). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experiments in our study were approved by the Institutional Animal Care and Use Committee of Yunnan Agricultural University (permission code: YAUACUC01; date of publication: 10 July 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang S, Xia P, Rehm M, et al. Autophagy and cell reprogramming. Cell Mol Life Sci 2015;72:1699-713. [Crossref] [PubMed]
- Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 2018;20:233-42. [Crossref] [PubMed]
- Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J 2017;36:1811-36. [Crossref] [PubMed]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2010;22:132-9. [Crossref] [PubMed]
- Funderburk SF, Wang QJ, Yue Z. Beclin 1-VPS34 complex – At the Crossroads of Autophagy and Beyond. Trends Cell Biol 2010;20:355-62. [Crossref] [PubMed]
- Xu DW, Zhang GQ, Wang ZW, et al. Autophagy in tumorigenesis and cancer treatment. Asian Pac J Cancer Prev 2015;16:2167-75. [Crossref] [PubMed]
- Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012;198:219-33. [Crossref] [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528. [Crossref] [PubMed]
- Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell 2017;170:1062-78. [Crossref] [PubMed]
- Sui X, Han W, Pan H. p53-induced autophagy and senescence. Oncotarget 2015;6:11723-4. [Crossref] [PubMed]
- Lu Z, Chen C, Wu Z, et al. A Dual Role of P53 in Regulating Colistin-Induced Autophagy in PC-12 Cells. Front Pharmacol 2017;8:768. [Crossref] [PubMed]
- Tasdemir E, Maiuri MC, Morselli E, et al. A dual role of p53 in the control of autophagy. Autophagy 2008;4:810-4. [Crossref] [PubMed]
- Feng Z, Zhang H, Levine AJ, et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A 2005;102:8204-9. [Crossref] [PubMed]
- Denisenko TV, Pivnyuk AD, Zhivotovsky B. p53-Autophagy-Metastasis Link. Cancers (Basel) 2018;10:148. [Crossref] [PubMed]
- Levine B, Abrams J. p53: The Janus of autophagy? Nature Cell Biology 2008;10:637-9. [Crossref] [PubMed]
- Tang J, Di J, Cao H, et al. p53-mediated autophagic regulation: A prospective strategy for cancer therapy. Cancer Lett 2015;363:101-7. [Crossref] [PubMed]
- Shen Y, Xu K, Yuan Z, et al. Efficient generation of P53 biallelic knockoutDiannanminiature pigs via TALENs and somatic cell nuclear transfer. J Transl Med 2017;15:224. [Crossref] [PubMed]
- Zhu W, Qu H, Xu K, et al. Differences in the starvation-induced autophagy response in MDA-MB-231 and MCF-7 breast cancer cells. Anim Cells Syst (Seoul) 2017;21:190-8. [Crossref] [PubMed]
- Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012;8:445-544. [Crossref] [PubMed]
- Gupta NA, Kolachala VL, Jiang R, et al. Mitigation of autophagy ameliorates hepatocellular damage following ischemia-reperfusion injury in murine steatotic liver. Am J Physiol Gastrointest Liver Physiol 2014;307:G1088-99. [Crossref] [PubMed]
- Paglin S, Hollister T, Delohery T, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res 2001;61:439-44. [PubMed]
- Choi JC, Worman HJ. Reactivation of autophagy ameliorates LMNA cardiomyopathy. Autophagy 2013;9:110-1. [Crossref] [PubMed]
- Li X, Wu C, Chen N, et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016;7:33440-50. [PubMed]
- Miki Y, Tanji K, Mori F, et al. Autophagy mediators (FOXO1, SESN3 and TSC2) in Lewy body disease and aging. Neurosci Lett 2018;684:35-41. [Crossref] [PubMed]
- Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 2018;19:349-64. [Crossref] [PubMed]
- White E. Autophagy and p53. Cold Spring Harb Perspect Med 2016;6:a026120. [Crossref] [PubMed]
- Gauster M, Maninger S, Siwetz M, et al. Downregulation of p53 drives autophagy during human trophoblast differentiation. Cell Mol Life Sci 2018;75:1839-55. [Crossref] [PubMed]
- Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008;10:676-87. [Crossref] [PubMed]
- Alers S, Löffler AS, Wesselborg S, et al. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 2012;32:2-11. [Crossref] [PubMed]
- Park JM, Seo M, Jung CH, et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 2018;14:584-97. [Crossref] [PubMed]
- Soto-Burgos J, Zhuang X, Jiang L, et al. Dynamics of autophagosome formation. Plant Physiol 2018;176:219-29. [Crossref] [PubMed]
- Torii S, Yoshida T, Arakawa S, et al. Identification of PPM1D as an essential Ulk1 phosphatase for genotoxic stress-induced autophagy. EMBO Rep 2016;17:1552-64. [Crossref] [PubMed]


