
Uncovering the potential miRNAs and mRNAs in follicular variant of papillary thyroid carcinoma in the Cancer Genome Atlas database
Introduction
Thyroid carcinoma, originated from parafollicular or follicular thyroid cells, possesses the highest incidence in the endocrine neoplasias (1). Papillary thyroid carcinoma, medullary thyroid carcinoma, anaplastic thyroid carcinoma, follicular thyroid carcinoma and poorly differentiated thyroid carcinoma is five types of thyroid carcinoma (2). The follicular variant of papillary thyroid carcinoma, a heterogeneous tumor, is the second most common morphologic subtype of papillary thyroid carcinomas (3,4). The follicular variant of papillary thyroid carcinoma has a follicular architecture that is lined by cells with nuclear features of papillary thyroid carcinoma. The follicular variant of papillary thyroid carcinoma includes the encapsulated and infiltrating variants (5). It is hypothesized that the follicular variant of papillary thyroid carcinoma could reflect aggressive parameters including bilateral lesions, vascular invasion, extrathyroid extension and distant metastasis (such as pulmonary metastases and bone metastases) at the time of diagnosis (3,6-9). The prognosis of follicular variant of papillary thyroid carcinoma is favorable and the 10- and 15-year overall survival is 93% and 89%, respectively (10).
MicroRNAs (miRNAs) are small and non-coding single-chained RNAs, which targets mRNAs and causing their translation or degradation (11). Several evidences indicate that miRNAs have crucial functions in adjusting biological behaviors including cell proliferation, apoptosis and differentiation (12,13). It is reported that hsa-mir-146-5p, -146b-3p, -222, -222-5, -221, -551b, -375, 99b-3p and 181-2-3p are deregulated in follicular variant of papillary thyroid carcinoma. Furthermore, hsa-mir-181a-2-3p and hsa-mir-99b-3p are related to the adverse outcome in patients with follicular variant of papillary thyroid carcinoma (14). DNA methylation can modify mRNA expression patterns and aberrant hypermethylation is associated with inactivation of tumor-related mRNAs (15,16). The methylation pattern of NIS, TSHR, SLC26A4, TIMP3, RARβ2 and RASSF1A has been found in papillary thyroid carcinoma (17-20).
In order to understand the pathology of the follicular variant of papillary thyroid carcinoma, we first performed the comprehensive analysis of miRNA, mRNA and DNA methylation profiling data from the Cancer Genome Atlas (TCGA) database. Differentially expressed miRNAs, mRNAs and methylated mRNAs were identified. Then, we performed the function enrichment analysis of differentially expressed target mRNAs of differentially expressed miRNAs. Last, we performed the real-time fluorescence quantitative polymerase chain reaction (QRT-PCR) to validate the expression of identified miRNAs and mRNAs.
Methods
Datasets
In this study, we downloaded the miRNA expression (miRNASeq BCGSC IlluminaHiSeq-miRNASeq), mRNA expression (RNASeqV2 UNC IlluminaHiseq_RNASeqV2) and DNA methylation data (Methyl JHU-USC HumanMethylation450) in tumor sample of 97 patients with follicular variant of papillary thyroid carcinoma from TCGA database (http://tcga-data.nci.nih.gov/) (April 2016).
Identification of differentially expressed miRNAs and mRNAs
Differentially expressed miRNAs and mRNAs were evaluated in the r-bioconductor package DESeq (21). The Limma package in R was applied to calculate P values by two-tailed Student’s t-test. MetaMA package in R was utilized to combine P values, and the false discovery rate was obtained from multiple comparisons using the Benjamini and Hochberg method (22). Those differentially expressed miRNAs and mRNAs were identified with the criterion of P<0.05.
Correlation analysis of differentially expressed miRNAs and mRNAs
The pairwise Pearson correlation coefficients between differentially expressed miRNAs and mRNAs were calculated, and P<0.05 was considered as statistical significance. Six miRNA-target prediction tools (miRWalk, miRanda, miRDB, RNA22, PICTAR2 and Targetscan) were used to predict target genes of differentially expressed miRNAs. Only those miRNA-target pairs which were predicted by more than four algorithms can be selected out. The miRNA-targets pairs verified by experiment in miRWalk database were also screened out. We selected the miRNA-target pairs with negative correlations to establish the miRNA-target regulatory network, which was visualized using Cytoscape software (23).
Functional analysis of target mRNAs of differentially expressed miRNAs
In order to understand the biological function of target mRNAs of differentially expressed miRNAs, we conducted Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis based on the online software GENECODIS (24). FDR <0.05 was set as the criterion for selecting significantly enriched GO and KEGG terms of target differentially expressed mRNAs.
Influence of DNA methylation on differentially expressed mRNAs
The COHCAP package in R (https://sourceforge.net/projects/cohcap/) (25) was used to screen the differential methylation sites between tumors and normal tissues that are more likely to regulate downstream gene expression. Methyl.cutoff =0.5, unmethyl.cutoff =0.3, delta.beta.cutoff =0.1 and false discovery rate <0.05 were considered as differentially methylated CpG sites. Based on the mRNA expression of the follicular variant of papillary thyroid carcinoma, we identified the aberrant DNA methylated CpG sites which affected corresponding mRNA expression.
QRT-PCR in vitro
Five patients diagnosed with follicular variant of papillary thyroid carcinoma and five normal individuals were enrolled in this study. Both the tumor and corresponding normal tissue samples were resected at the time of surgery, and immediately frozen in liquid nitrogen. All participating individuals provided informed consent with the approval of the ethics committee of General Hospital of Jinan Military Region of PLA. In addition, our study conforms to the provision and in accordance with the Helsinki Declaration.
Total RNA of the tissue samples was extracted using the TRIzol® Reagent (TIANGEN) according to the manufacturer’s protocols. Two µg RNA was applied to synthesize DNA by SuperScript® III Reverse Transcriptase (TIANGEN). Then real-time PCR was performed in an ABI 7300 real-time PCR system with SYBR® Green PCR Master Mix (Applied Biosystems). All reactions were carried out in triplicate. Relative gene expressions were analyzed by 2−ΔΔCt method.
Results
MiRNA and mRNA expression pattern
In this study, a total of 97 tissue samples were included in the present study, all of which with fully characterized miRNA profiles, mRNA profiles and DNA methylation data. There were 8 differentially expressed (4 up-regulated and 4 down-regulated) miRNAs and 973 differentially expressed (554 up-regulated and 419 down-regulated) mRNAs with P<0.05 between the follicular variant of papillary thyroid carcinomas and normal tissue. Eight identified differentially expressed miRNAs and top 10 up- and down-regulated differentially expressed mRNAs were listed in Table 1 and Table 2, respectively. The heat map of all differentially expressed miRNAs and top 50 differentially expressed mRNAs was showed in Figure 1 and Figure 2, respectively.
Table 1
| miRNA | log2Fold change | P value |
|---|---|---|
| Up-regulated miRNAs | ||
| hsa-mir-221 | 3.544169565 | 2.80E-04 |
| hsa-mir-222 | 3.21622386 | 8.37E-04 |
| hsa-mir-34a | 2.666407907 | 9.17E-03 |
| hsa-mir-1251 | 2.072575193 | 4.00E-02 |
| Down-regulated miRNAs | ||
| hsa-mir-138-2 | −0.982954891 | 4.47E-02 |
| hsa-mir-675 | −2.176635768 | 4.56E-02 |
| hsa-mir-138-1 | −0.972380058 | 4.79E-02 |
| hsa-mir-214 | −1.350097616 | 4.96E-02 |
Table 2
| Gene ID | Symbol | log2Fold change | P value |
|---|---|---|---|
| Up-regulated mRNAs | |||
| 401498 | TMEM215 | 9.972515 | 0.000169 |
| 170261 | ZCCHC12 | 8.728899 | 0.004099 |
| 6585 | SLIT1 | 8.139089 | 0.000174 |
| 222171 | PRR15 | 7.043451 | 0.000195 |
| 200879 | LIPH | 6.588005 | 3.60E-05 |
| 55220 | KLHDC8A | 6.559447 | 1.10E-06 |
| 2561 | GABRB2 | 6.261426 | 0.013721 |
| 10686 | CLDN16 | 5.924016 | 0.026352 |
| 120892 | LRRK2 | 5.57023 | 2.91E-05 |
| 51440 | HPCAL4 | 5.501627 | 0.032491 |
| Down-regulated mRNAs | |||
| 6366 | CCL21 | −5.06223 | 0.000305 |
| 90632 | C6orf176 | −4.21365 | 0.018373 |
| 221091 | LRRN4CL | −4.12688 | 1.69E-05 |
| 5649 | RELN | −4.02531 | 6.50E-09 |
| 347 | APOD | −3.94221 | 2.28E-07 |
| 1805 | DPT | −3.83603 | 3.31E-06 |
| 57528 | KCTD16 | −3.81913 | 4.87E-06 |
| 130497 | OSR1 | −3.81534 | 0.000239 |
| 125 | ADH1B | −3.5074 | 0.001008 |
| 10894 | LYVE1 | −3.48926 | 4.08E-07 |
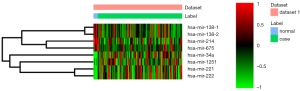
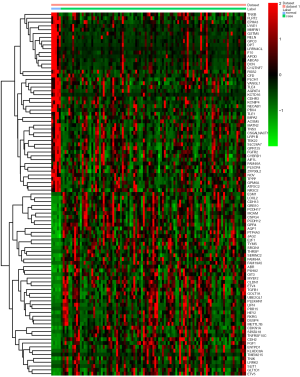
Correlations of differentially expressed miRNAs and mRNAs
In order to investigate the correlations between differentially expressed miRNAs and mRNAs, the correlation analysis was performed. Depending on the correlation analysis, 881 miRNA-mRNA pairs which were negatively correlated (P<0.05, r<−0.2) were identified. In the targeted analysis, 466 miRNA-mRNA pairs including 218 miRNA (up-regulation)-mRNA (down-regulated) pairs and 248 miRNA (down-regulation)-mRNA (up-regulated) pairs were screened out. The established regulatory network of miRNA-targeted mRNA with negative correlation was showed in Figure 3. The network was consisted of 6 differentially expressed (3 up-regulated and 3 down-regulated) miRNAs and 65 differentially expressed (25 up-regulated and 40 down-regulated) mRNAs. In addition, there were 71 nodes and 92 edges in the network. Interestingly, hsa-mir-222 (degree =33), hsa-mir-221 (degree =29), hsa-mir-214 (degree =13), hsa-mir-138-2 (degree =11) and hsa-mir-34a (degree =4) were miRNAs that regulated the most target mRNAs.
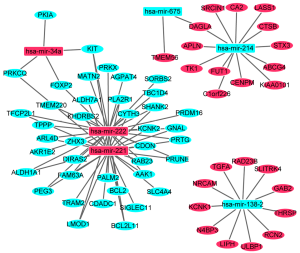
Functional analysis of target differentially expressed mRNAs of differentially expressed miRNAs
To understand the potential function of target differentially expressed mRNAs of differentially expressed miRNAs, we conducted GO and KEGG pathway analysis. The results demonstrated that these targeted differentially expressed mRNAs were most significantly enriched in the biological processes of signal transduction, angiogenesis and cell adhesion. Additionally, focal adhesion, pathways in cancer, gap junction, cytokine-cytokine receptor interaction, the MAPK signaling pathway, p53 signaling pathway and ECM-receptor interaction were the significantly enriched signaling pathways of targeted differentially expressed mRNAs. Top 15 significantly enriched GO terms (biological process) and KEGG terms of targeted differentially expressed mRNAs were listed in Table 3 and Table 4, respectively.
Table 3
| GO ID | GO term | Number of genes | False discovery rate |
|---|---|---|---|
| 0007165 | Signal transduction | 42 | 1.09E-10 |
| 0001525 | Angiogenesis | 16 | 8.12E-10 |
| 0007155 | Cell adhesion | 27 | 9.64E-10 |
| 0030168 | Platelet activation | 17 | 1.42E-08 |
| 0007275 | Multicellular organismal development | 33 | 1.65E-08 |
| 0030324 | Lung development | 10 | 1.79E-07 |
| 0007596 | Blood coagulation | 15 | 2.10E-07 |
| 0001701 | In utero embryonic development | 6 | 3.30E-07 |
| 0042493 | Response to drug | 4 | 3.62E-07 |
| 0032026 | Response to magnesium ion | 4 | 3.62E-07 |
| 0032355 | Response to estradiol stimulus | 4 | 3.62E-07 |
| 0010544 | Negative regulation of platelet activation | 4 | 3.62E-07 |
| 0001666 | Response to hypoxia | 6 | 1.17E-06 |
| 0030335 | Positive regulation of cell migration | 4 | 1.24E-06 |
| 0032355 | Response to estradiol stimulus | 9 | 5.20E-06 |
Table 4
| KEGG ID | KEGG items | Number of genes | False discovery rate |
|---|---|---|---|
| Hsa04510 | Focal adhesion | 15 | 1.01E-07 |
| Hsa04810 | Regulation of actin cytoskeleton | 13 | 3.69E-06 |
| Hsa05215 | Prostate cancer | 7 | 3.72E-06 |
| Hsa05200 | Pathways in cancer | 16 | 4.37E-06 |
| Hsa05218 | Melanoma | 6 | 6.60E-06 |
| Hsa05214 | Glioma | 6 | 3.66E-05 |
| Hsa04360 | Axon guidance | 9 | 4.27E-05 |
| Hsa04540 | Gap junction | 4 | 4.32E-05 |
| Hsa04060 | Cytokine-cytokine receptor interaction | 12 | 5.64E-05 |
| Hsa04010 | MAPK signaling pathway | 3 | 8.88E-05 |
| Hsa04115 | p53 signaling pathway | 3 | 8.88E-05 |
| Hsa04512 | ECM-receptor interaction | 7 | 9.65E-05 |
| Hsa04210 | Apoptosis | 7 | 9.73E-05 |
| Hsa04020 | Calcium signaling pathway | 9 | 1.97E-04 |
| Hsa05221 | Acute myeloid leukemia | 5 | 4.81E-04 |
Influence of DNA methylation on differentially expressed mRNAs
Generally, the methylation of CpG islands in the promoter regions results in the mRNA silencing. There were 186 differentially methylated CpG sites with FDR <0.05, which involved 146 mRNAs. However, all these differentially methylated mRNAs were not differentially expressed mRNAs identified above. Therefore, we mainly focused on the study of differentially expressed miRNAs and mRNAs in the follicular variant of papillary thyroid carcinoma.
qRT-PCR
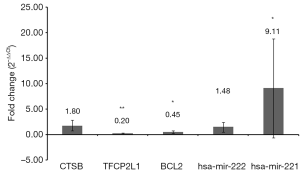
Five pairs of follicular variants of papillary thyroid carcinoma and adjacent normal tissues were used to validate the expression of identified differentially expressed miRNAs and mRNAs. Based on the above analysis, 2 differentially expressed miRNAs (hsa-mir-222 and hsa-mir-221) and 3 differentially expressed mRNAs (CTSB, TFCP2L1 and BCL2) were randomly selected for validation (Figure 4). The qRT-PCR results demonstrated that hsa-mir-221 was significantly up-regulated (P<0.05), TFCP2L1 and BCL2 were remarkably down-regulated (P<0.01 and P<0.05, respectively). Hsa-mir-222 and CTSB were up-regulated without significance. All these results were consistent data with the integrated analysis.
Discussion
Identification of changes of miRNAs and mRNAs in different levels occurring in tumor development is an important step in fighting against the follicular variant of follicular variant of papillary thyroid carcinoma. In this study, we integrated miRNA expression, mRNA expression data and DNA methylation profiles of follicular variant of papillary thyroid carcinoma to find valuable miRNAs and mRNAs which were highly associated with the follicular variant of papillary thyroid carcinoma. In the network of miRNA-mRNA with the negative correlation, we obtained 5 differentially expressed miRNAs including hsa-mir-222 (up-regulation), hsa-mir-221 (up-regulation), hsa-mir-34a (up-regulation), hsa-mir-214 (down-regulation) and hsa-mir-138-2 (down-regulation) that regulated the most target mRNAs.
Hsa-mir-222 is a cancer-related miRNA and can promote the proliferation of cancer cell (26). It is reported that up-regulated expression of hsa-mir-222 is characteristic of papillary thyroid carcinoma (27). In addition, over expression of hsa-mir-222 is significantly associated with tumor aggression, central lymph node metastases, extrathyroidal invasion and recurrence, which were considered as the independent predictor of papillary thyroid carcinoma prognosis (28-31). Visone et al. found that hsa-mir-221 was excessively secreted in papillary thyroid carcinoma (32). Over expression of hsa-mir-221 is remarkably correlated with tumor aggression and clinic-pathological characteristic of papillary thyroid carcinoma (28,29). It has been demonstrated that hsa-mir-221 could be a potential prognostic biomarker for the recurrence in papillary thyroid carcinoma (33). In this study, we found the relationship between hsa-mir-222, hsa-mir-221 and follicular variant of papillary thyroid carcinoma. In addition, we found that 7 down-regulated differentially expressed mRNAs including 3 cell apoptosis related mRNAs (BCL2, BCL2L11 and PEG3), ALDH1A1, PLA2R1, TFCP2L1 and RAB23 were commonly regulated by hsa-mir-222 and hsa-mir-221.
BCL2, apoptosis regulator (BCL2), an antiapoptotic protein, is involved in biological processes of cell survival and blocking of apoptosis (34,35). It is found that the over expression of BCL2 is associated with aggressiveness of papillary thyroid carcinoma (36). BCL2 like 11 (BCL2L11) is a pro-apoptotic gene and involved in controlling cell death. Bim protein, encoded by BCL2L11, has been regarded as functionally relevant in rat follicular thyroid cells (37). Paternally expressed 3 (PEG3) is associated with cell death and apoptosis. PEG3 encodes the tumor suppressor and is down-regulated in several cancer types, such as glioma, gynecologic cancer, ovarian cancer and invasive cervical cancer (38-41). This suggested that cell apoptosis plays an important role in the process of follicular variant of papillary thyroid carcinoma.
It is reported that aldehyde dehydrogenase 1 family member A1 (ALDH1A1) is down-regulated in papillary thyroid carcinoma (42). In addition, the expression of ALDH1A1 is significantly associated with factors (such as lymph node metastasis and extrathyroidal extension) that lead to poor prognosis of papillary thyroid carcinoma (43). Finn et al. found that phospholipase A2 receptor 1 (PLA2R1) was down-regulated in malignant papillary thyroid carcinoma, and appears to suppress tumorigenesis by activating the tyrosine kinase JAK2 (44). It is noted that PLA2R1 is associated with the prognosis of papillary thyroid carcinoma (45). Kim et al. found that lower expression of TFCP2L1 was down-regulated in papillary thyroid carcinoma compared to normal thyroid sample (46). It is worth mentioning that the expression of TFCP2L1 is detected in the follicular variant of papillary thyroid carcinoma (47). RAB23, member RAS oncogene family (RAB23) is a protein that belongs to the Rab family of GTPases. It has been demonstrated that over expression of RAB23 promotes proliferation, migration, invasion and metastasis of tumor cell (48,49). It is noted that the expression of RAB23 is decreased in the follicular variant of papillary thyroid carcinoma compared to the benign follicular adenoma (50).
It is reported that hsa-mir-34a is up-regulated in tissues and cell lines of papillary thyroid carcinoma (51,52). Moreover, the expression level of hsa-mir-34a is associated with tumor invasion and showed the potential diagnostic value of papillary thyroid carcinoma (53). Herein, we found up-regulated expression of hsa-mir-34a in the follicular variant of papillary thyroid carcinoma. Furthermore, protein kinase C theta (PRKCQ) and prune exopolyphosphatase (PRUNE) were two of targets of hsa-mir-34a. PRKCQ is related to immune response and down-regulated in the brain metastatic of papillary thyroid carcinoma (54). It is found that the expression of PRUNE is detected in anaplastic thyroid cancer and promotes invasion, migration and metastasis of anaplastic thyroid cancer cell (55).
Hsa-mir-214 is significantly down-regulated in tissues and cells of papillary thyroid carcinoma compared with normal, which was remarkably associated with tumor size, TNM stage and lymph node metastasis. In this study, we found that the expression of hsa-mir-214 is down-regulated in the follicular variant of papillary thyroid carcinoma. Moreover, Thymidine kinase 1 (TK1) was one of targets of hsa-mir-214. TK1 is involved in DNA repair and internationally recognized as the marker of abnormal cell proliferation. It is noted that up-regulated expression of TK1 is closely associated with active tumor growth (56). Furthermore, serological TK1 could be an important marker for the early risk of malignancy development (57).
It has been demonstrated that hsa-mir-138-2 has a diagnostic value for papillary thyroid carcinoma (53). In the present study, we found that the expression of hsa-mir-138-2 was decreased in the follicular variant of papillary thyroid carcinoma. In addition, cathepsin (CTSB) was regulated by hsa-mir-138-2. CTSB, a lysosome enzyme, is over expressed in tumor endothelial and epithelial cells. It is found that CTSB is significantly up-regulated in the tumor tissues of thyroid cancer.
According to the KEGG analysis, we found that focal adhesion, MAPK signaling pathway and p53 signaling pathway were three significantly enriched signaling pathways of targeted differentially expressed mRNAs of differentially expressed miRNAs. The focal adhesion signaling pathway is important in maintaining cellular physiology. It is reported that the focal adhesion signaling pathway play a crucial role in the pathogenesis of renal cell carcinoma (58). It is found that constitutive activation of MAPK signaling pathway plays an important role in thyroid carcinoma tumorigenesis (59). Moreover, MAPK signaling pathway is a promising therapeutic target for thyroid cancer (60-62). P53, the tumor suppressor, regulates a lot of biological processes including DNA damage, cellular senescence and apoptosis (63,64). It is reported that p53 mutation is recently detected in about 40% of papillary thyroid carcinoma (65).
Conclusions
In summary, our integrated analysis of the TCGA data led to a number of differentially expressed miRNAs, mRNAs and methylated mRNAs. The epigenetic modifications via five miRNAs (hsa-mir-222, hsa-mir-221, hsa-mir-34a, hsa-mir-214 and hsa-mir-138-2) for BCL2, BCL2L11 and PEG3, ALDH1A1, PLA2R1, TFCP2L1, RAB23, TK1 and CTSB may be involved in tumorigenesis of follicular variant of papillary thyroid carcinoma. The study of epigenetic alterations between these miRNAs and mRNAs is of value to investigate the pathogenesis of follicular variant of papillary thyroid carcinoma. However, there are limitations to our study. Firstly, the sample in the QRT-PCR is small and larger numbers of tumor tissues are needed for validation; secondly, the deeper mechanism study of the disease such as the animal model and cell culture are also needed.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All participating individuals provided informed consent with the approval of the ethics committee of General Hospital of Jinan Military Region of PLA. In addition, our study conforms to the provision and in accordance with the Helsinki Declaration.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Carling T, Udelsman R. Thyroid cancer. Annu Rev Med 2014;65:125-37. [Crossref] [PubMed]
- Carcangiu ML, Zampi G, Pupi A, et al. Papillary carcinoma of the thyroid. A clinicopathologic study of 241 cases treated at the University of Florence, Italy. Cancer 1985;55:805-28. [Crossref] [PubMed]
- Lam AK, Lo CY, Lam KS. Papillary carcinoma of thyroid: A 30-yr clinicopathological review of the histological variants. Endocr Pathol 2005;16:323-30. [Crossref] [PubMed]
- Shi X, Liu R, Basolo F, et al. Differential Clinicopathological Risk and Prognosis of Major Papillary Thyroid Cancer Variants. J Clin Endocrinol Metab 2016;101:264-74. [Crossref] [PubMed]
- Hagag P, Hod N, Kummer E, et al. Follicular variant of papillary thyroid carcinoma: clinical-pathological characterization and long-term follow-up. Cancer J 2006;12:275-82. [Crossref] [PubMed]
- Chang HY, Lin JD, Chou SC, et al. Clinical presentations and outcomes of surgical treatment of follicular variant of the papillary thyroid carcinomas. Jpn J Clin Oncol 2006;36:688-93. [Crossref] [PubMed]
- Cardenas MG, Kini S, Wisgerhof M. Two patients with highly aggressive macrofollicular variant of papillary thyroid carcinoma. Thyroid 2009;19:413-6. [Crossref] [PubMed]
- Liu L, Venkataraman G, Salhadar A. Follicular variant of papillary thyroid carcinoma with unusual late metastasis to the mandible and the scapula. Pathol Int 2007;57:296-8. [Crossref] [PubMed]
- Yu XM, Schneider DF, Leverson G, et al. Follicular variant of papillary thyroid carcinoma is a unique clinical entity: a population-based study of 10,740 cases. Thyroid 2013;23:1263. [Crossref] [PubMed]
- Jazdzewski K, Liyanarachchi S, Swierniak M, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A 2009;106:1502-5. [Crossref] [PubMed]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007;449:682-8. [Crossref] [PubMed]
- Vriens MR, Weng J, Suh I, et al. MicroRNA expression profiling is a potential diagnostic tool for thyroid cancer. Cancer 2012;118:3426-32. [Crossref] [PubMed]
- Dettmer M, Perren A, Moch H, et al. Comprehensive MicroRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid 2013;23:1383-9. [Crossref] [PubMed]
- Wu H, Sun YE. Epigenetic regulation of stem cell differentiation. Pediatr Res 2006;59:21R-5R. [Crossref] [PubMed]
- Issa JP, Baylin SB, Herman JG. DNA methylation changes in hematologic malignancies: biologic and clinical implications. Leukemia 1997;11:S7-11. [PubMed]
- Xing M, Usadel H, Cohen Y, et al. Methylation of the thyroid-stimulating hormone receptor gene in epithelial thyroid tumors: a marker of malignancy and a cause of gene silencing. Cancer Res 2003;63:2316-21. [PubMed]
- Xing M, Tokumaru Y, Wu G, et al. Hypermethylation of the Pendred syndrome gene SLC26A4 is an early event in thyroid tumorigenesis. Cancer Res 2003;63:2312-5. [PubMed]
- Schagdarsurengin U, Gimm O, Hoang-Vu C, et al. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res 2002;62:3698-701. [PubMed]
- Brait M, Loyo M, Rosenbaum E, et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARβ2 and RASSF1A in thyroid cancer. Epigenetics 2012;7:710-9. [Crossref] [PubMed]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser 1995;57:289-300.
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res 2012;40:W478-83. [Crossref] [PubMed]
- Warden CD, Lee H, Tompkins JD, et al. COHCAP: an integrative genomic pipeline for single-nucleotide resolution DNA methylation analysis. Nucleic Acids Res 2013;41:e117. [Crossref] [PubMed]
- Li N, Tang B, Zhu ED, et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett 2012;586:722-8. [Crossref] [PubMed]
- Titov SE, Ivanov MK, Karpinskaya EV, et al. miRNA profiling, detection of BRAF V600E mutation and RET-PTC1 translocation in patients from Novosibirsk oblast (Russia) with different types of thyroid tumors. BMC Cancer 2016;16:201. [Crossref] [PubMed]
- Nikiforova MN, Tseng GC, Steward D, et al. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 2008;93:1600-8. [Crossref] [PubMed]
- Lee JC, Ting ZJ, Clifton-Bligh RJ, et al. MicroRNA-222 and MicroRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 2013;119:4358-65. [Crossref] [PubMed]
- Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid 2010;20:489-94. [Crossref] [PubMed]
- Han PA, Kim HS, Cho S, et al. Association of BRAF V600E Mutation and MicroRNA Expression with Central Lymph Node Metastases in Papillary Thyroid Cancer: A Prospective Study from Four Endocrine Surgery Centers. Thyroid 2016;26:532-42. [Crossref] [PubMed]
- Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer 2007;14:791-8. [Crossref] [PubMed]
- Dai L, Wang Y, Chen L, et al. MiR-221, a potential prognostic biomarker for recurrence in papillary thyroid cancer. World J Surg Oncol 2017;15:11. [Crossref] [PubMed]
- Mitsiades CS, Patrick H, Vassiliki K, et al. Bcl-2 overexpression in thyroid carcinoma cells increases sensitivity to Bcl-2 homology 3 domain inhibition. J Clin Endocrinol Metab 2007;92:4845-52. [Crossref] [PubMed]
- Cleland MM, Norris KL, Karbowski M, et al. Bcl-2 family interaction with the mitochondrial morphogenesis machinery. Cell Death Differ 2011;18:235-47. [Crossref] [PubMed]
- Siironen P, Nordling S, Louhimo J, et al. Immunohistochemical Expression of Bcl-2, Ki-67, and p21 in Patients with Papillary Thyroid Cancer. Tumour Biol 2005;26:50-6. [Crossref] [PubMed]
- Karger S, Weidinger C, Krause K, et al. FOXO3a: a novel player in thyroid carcinogenesis? Endocr Relat Cancer 2009;16:189-99. [Crossref] [PubMed]
- Chen MY, Liao WS, Lu Z, et al. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer 2011;117:4424-38. [Crossref] [PubMed]
- Dowdy SC, Gostout BS, Shridhar V, et al. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol Oncol 2005;99:126-34. [Crossref] [PubMed]
- Jiang X, Yu Y, Yang HW, et al. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J Biol Chem 2010;285:8472-80. [Crossref] [PubMed]
- Nye MD, Hoyo C, Huang Z, et al. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One 2013;8:e56325. [Crossref] [PubMed]
- Beltrami CM, Dos Reis MB, Barros-Filho MC, et al. Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas. Clin Epigenetics 2017;9:45. [Crossref] [PubMed]
- Xing Y, Luo DY, Long MY, et al. High ALDH1A1 expression correlates with poor survival in papillary thyroid carcinoma. World J Surg Oncol 2014;12:29. [Crossref] [PubMed]
- Finn SP, Smyth P, Cahill S, et al. Expression microarray analysis of papillary thyroid carcinoma and benign thyroid tissue: emphasis on the follicular variant and potential markers of malignancy. Virchows Arch 2007;450:249-60. [Crossref] [PubMed]
- Brennan K, Holsinger C, Dosiou C, et al. Development of prognostic signatures for intermediate-risk papillary thyroid cancer. BMC Cancer 2016;16:736. [Crossref] [PubMed]
- Kim HS, Kim DH, Kim JY, et al. Microarray analysis of papillary thyroid cancers in Korean. Korean J Intern Med 2010;25:399-407. [Crossref] [PubMed]
- Schulten HJ, Al-Mansouri Z, Baghallab I, et al. Comparison of microarray expression profiles between follicular variant of papillary thyroid carcinomas and follicular adenomas of the thyroid. BMC Genomics 2015;16:S7. [Crossref] [PubMed]
- Jian Q, Miao Y, Tang L, et al. Rab23 promotes squamous cell carcinoma cell migration and invasion via integrin β1/Rac1 pathway. Oncotarget 2016;7:5342-52. [Crossref] [PubMed]
- Jiang Y, Han Y, Sun C, et al. Rab23 is overexpressed in human bladder cancer and promotes cancer cell proliferation and invasion. Tumour Biol 2016;37:8131-8. [Crossref] [PubMed]
- Denning KM, Smyth PC, Cahill SF, et al. A molecular expression signature distinguishing follicular lesions in thyroid carcinoma using preamplification RT-PCR in archival samples. Mod Pathol 2007;20:1095-102. [Crossref] [PubMed]
- Ma Y, Qin H, Cui Y. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun 2013;441:958-63. [Crossref] [PubMed]
- Lu X, Chen Z, Liang H, et al. Thyroid hormone inhibits TGFβ1 induced renal tubular epithelial to mesenchymal transition by increasing miR34a expression. Cell Signal 2013;25:1949-54. [Crossref] [PubMed]
- Cong D, He M, Chen S, et al. Expression profiles of pivotal microRNAs and targets in thyroid papillary carcinoma: an analysis of The Cancer Genome Atlas. Onco Targets Ther 2015;8:2271-7. [PubMed]
- Lim EL, Trinh DL, Scott DW, et al. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol 2015;16:18. [Crossref] [PubMed]
- Nambu J, Kobayashi T, Hashimoto M, et al. h-prune affects anaplastic thyroid cancer invasion and metastasis. Oncol Rep 2016;35:3445-52. [Crossref] [PubMed]
- Carlsson L, Larsson A, Lindman H. Elevated levels of thymidine kinase 1 peptide in serum from patients with breast cancer. Ups J Med Sci 2009;114:116-20. [Crossref] [PubMed]
- Chen Z, Zhou H, Li S, et al. Serological thymidine kinase 1 (STK1) indicates an elevated risk for the development of malignant tumours. Anticancer Res 2008;28:3897-907. [PubMed]
- Wala SJ, Karamchandani JR, Saleeb R, et al. An integrated genomic analysis of papillary renal cell carcinoma type 1 uncovers the role of focal adhesion and extracellular matrix pathways. Mol Oncol 2015;9:1667-77. [Crossref] [PubMed]
- Shen CT, Qiu ZL, Song HJ, et al. miRNA-106a directly targeting RARB associates with the expression of Na + /I − symporter in thyroid cancer by regulating MAPK signaling pathway. J Exp Clin Cancer Res 2016;35:101. [Crossref] [PubMed]
- Petrulea MS, Plantinga TS, Smit JW, et al. PI3K/Akt/mTOR: A promising therapeutic target for non-medullary thyroid carcinoma. Cancer Treat Rev 2015;41:707-13. [Crossref] [PubMed]
- Manfredi GI, Dicitore A, Gaudenzi G, et al. PI3K/Akt/mTOR signaling in medullary thyroid cancer: a promising molecular target for cancer therapy. Endocrine 2015;48:363-70. [Crossref] [PubMed]
- Milosevic Z, Pesic M, Stankovic T, et al. Targeting RAS-MAPK-ERK and PI3K-AKT-mTOR signal transduction pathways to chemosensitize anaplastic thyroid carcinoma. Transl Res 2014;164:411-23. [Crossref] [PubMed]
- Chen L, Rousseau RF, Middleton SA, et al. Pre-clinical evaluation of the MDM2-p53 antagonist RG7388 alone and in combination with chemotherapy in neuroblastoma. Oncotarget 2015;6:10207-21. [PubMed]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009;137:413-31. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676-90. [Crossref] [PubMed]


