
New approaches to model glioblastoma in vitro using brain organoids: implications for precision oncology
The urgent need for better therapies for glioblastoma
Glioblastoma is an aggressive form of primary brain cancer for which new treatment approaches are desperately needed. To achieve this goal, better experimental approaches to study human glioblastoma are required; and in particular, methods that recapitulate the native tumor microenvironment will be most valuable. In accordance with this need, Howard Fine and co-workers have developed a new experimental model, termed GLICO (1), which should facilitate a better understanding of the biology of the disease, and importantly, enable more reliable testing of potential new therapies.
Currently, standard therapy for glioblastoma involves surgery to achieve maximal safe resection, combined with radiotherapy and temozolomide chemotherapy. However, despite this multi-modal approach, tumor recurrence is almost inevitable, after which no treatments that effectively prolong survival exist (2,3). Median survival time from diagnosis is only 15 months, and the 5-year relative survival (just 4.6% at 5 years) has remained stable over the last three decades. Considering this dismal situation, there is intense interest in developing new treatments for glioblastoma. So far, however, those treatments that appeared initially effective against cultured cell lines and mouse tumor models have generally failed in the clinic (4). Possibly, more sophisticated experimental systems, such as that developed by Linkous et al. (1), will allow for more accurate prediction of the therapeutic efficacy of potential new treatments. And because this system mimics the native glioblastoma microenvironment, it may also facilitate the development of novel agents that specifically target non-malignant aspects of the tumor microenvironment, especially the extracellular matrix (ECM) and the neuronal and glial compartments.
GLICO: a new brain organoid model to grow patient-derived glioma stem cells (GSCs)
The model developed by Linkous et al. (1) involves the growth of patient-derived glioblastoma cells within cerebral organoids, which are complex, multicellular structures that mimic the architecture of a developing human brain (5) (Figure 1). This model was termed GLICO, for cerebral organoid glioma. The glioblastoma cells used to create GLICOs were cultured from patient tumor tissue under defined (serum-free) conditions that promote the maintenance of a stem cell phenotype, and are referred to as GSCs. The cerebral organoids were grown from human embryonic or induced pluripotent stem (iPS) cells, under conditions that promote 3D expansion of neuroectoderm, as previously described (5). When GSCs were co-cultured with fully formed cerebral organoids, the GSCs invaded and established tumors within the organoids with remarkable efficiency (100% tumor take rate), demonstrating the inherently invasive nature of glioblastoma cells, and the preservation of this capacity achieved by culturing cells under GSC conditions.
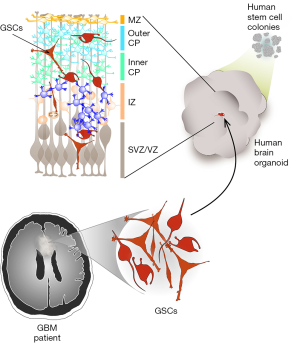
Morphologically, the growth of GSCs within cerebral organoids closely resembled human glioblastoma tumors, characterized by a hypercellular bulk tumor with a leading edge of GSCs infiltrating the surrounding normal (organoid) tissue, as well as marked necrosis. As expected, the GSCs were highly proliferative within the organoids, as detected by Ki67 immunostaining, incorporation of the thymidine analog EdU, and tracking the growth trajectory of cells over time. Less expected, but highly intriguing, was the observation of networks of microtubes connecting individual GSCs to each other, and GSCs to normal neurons in the cerebral organoid. These connections involved cytoplasmic fusion between individual cells, mediated by connexin-43-positive gap junctions and desmosomal junctions. Such microtubes have also recently been reported in human glioblastoma tumors, where they are thought to facilitate multicellular communication between cells, and to provide routes for invasion (6). The detection of these microtubes in the GLICO model provides evidence that the cancer cells have adopted behaviors and phenotypes in this system that closely mimic those of native tumors. GSCs grown within the GLICO system also maintained key genetic and signaling components of the source tumor, including amplification of the EGFR gene and patterns of receptor tyrosine kinase phosphorylation.
To demonstrate the practical value of their model in pre-clinical research, Linkous et al. compared the sensitivity of GSCs to chemotherapy or radiation when grown as GLICOs, compared to standard 2D tissue culture. GSCs were markedly more resistant to temozolomide, bis-chloroethylnitrosourea and ionizing radiation when grown within cerebral organoids, suggesting that this system effectively models the protective effect of the tumor microenvironment on glioblastoma cell survival. Thus, using the GLICO model to screen for new therapeutic agents may enable better predictions of their clinical effectiveness.
Other brain organoid models to study interactions between tumor cells and the surrounding healthy brain microenvironment
The last few years have shown significant progress in the development of assays to study glioblastoma cancer cells in 3D organoid models, and their interaction with ‘healthy’ brain tissue (3). These advances started in 2016 with the first description of glioblastoma patient-derived organoids created from tumor cells (CD133+) isolated from biopsies (7). These ‘tumoroids’ maintained key aspects of the source tumor, including regional heterogeneity and hypoxia gradients, and demonstrated a high tumorigenic capacity in patient-derived xenograft (PDX) models. Two years later, da Silva et al. progressed this approach to develop a co-culture model in which tumoroids were put in contact with early-stage brain organoids (8). Under these conditions, the authors were able to show an efficient and instant fusion between the tumoroid cell aggregates and the organoids, followed by rapid (~16 hours) spontaneous infiltration of tumor cells into the healthy brain organoid This report thus showed that studies of interactions between patient-derived cancer cells and healthy brain tissue were possible, but with a caveat. Instead of the multi-layered, mature human cerebral organoids used in the study by Linkous et al., this earlier study used mouse cells to create a primitive neuroepithelial structure with inside-out topology (i.e., no lumen and with the apical pole of cells exposed to the medium), which may at least partly explain the extremely fast fusion and infiltration of cancer cells.
Co-culture systems, including the one described above (8) and the study by Linkous et al. (1), present distinct advantages including ease of labeling (patient-derived) tumor and non-tumor cells with different colors for tracing, and analysis of tumor cell invasion within ‘healthy’ brain tissue. However, a critical limitation of co-culture approaches is an inability to model the interactions between tumor cells and the healthy microenvironment when tumor cells first arise. To circumvent this problem and study tumor growth at very early stages, the Knoblich lab in 2018 used an electroporation approach in combination with CRISPR/Cas9 to express oncogenes within cerebral organoids, resulting in spontaneous formation of tumor cells (9). This ‘neoplastic cerebral organoid’ (neoCOR) system efficiently models glioblastoma initiation, growth and invasion within a human brain microenvironment. At the same time, Ogawa et al. targeted the human TP53 locus using CRISPR/Cas9 to integrate an inducible RAS expression cassette to block TP53 activity within established human brain organoids (10), offering the extra advantage of inducing tumor formation even at very late stages of brain organoid maturation. Although both reports show several advantages, their models have the primary limitation of genetic manipulation, which can be only be performed on a few genes that are known to be molecular drivers of glioblastoma, and which are not necessarily representative of the heterogeneity of this cancer (3).
In addition to co-culture systems and genetic models of glioblastoma development, bio-printing is emerging as another relevant approach to model the glioblastoma microenvironment. This year, Yi et al. (11) reported the use of decellularized ECM from (pig) brain tissue as a bio-ink to create glioblastoma tumors containing patient-derived tumor cells, vascular endothelial cells and natural brain ECM. Of note, this bio-printed organ-on-a-chip platform reproduced clinically observed patient-specific responses to radiation and temozolomide. It is also very fast, requiring only 2–5 days to generate. However, this system lacks the high-order level of self-organization that occurs using current protocols for brain organoid culture (5).
The GLICO model, and many of the other complex systems discussed above, are limited by low/medium throughput capabilities and have only been validated using a small number of patient-derived samples [i.e., less than 10 (1,9,11)]. Moreover, development of most of these requires a significant period (generally 1–2 months) which may exceed a clinically relevant time frame (12). Considering the aggressiveness of this type of brain cancer and the modest extension of survival after standard therapy, this limited throughput capacity and extended timeframe constitute critical disadvantages that need to be addressed in the near future. If successful, this will leverage current efforts to build-up live tumor banks for access to patient-derived clinical samples for validation and reproducibility testing of these assays, which will be critical for their broad preclinical acceptance.
It is also important to emphasize that there are still key discrepancies between human tissues and the structures developed within these organoid models. One particular issue is the lack of vascularization, only overcome by transplanting organoids into the brains of mice (13), although a recent report has now shown (for a single patient specimen) that iPS cell-derived endothelial cells can be incorporated within brain organoids forming a blood vessel-like structure (14). The additional incorporation of immune cells and blood brain barrier (BBB) function [see for example (15,16)] as well as glia (17) and microglia (18,19) into glioblastoma-organoid models, will be exciting areas of research in the near future.
Application of brain organoid models in precision oncology
In glioblastoma, the high unmet clinical need together with a market size that is significant on a global level may justify interest and investment in models where patient-derived glioblastoma cells are hosted by mini-brains in vitro. Given that the model described by Linkous et al. can recreate the inherent therapeutic resistance of tumor cells cossetted in a supportive tumor microenvironment (1), precision oncology and cancer drug development are two major areas where this approach could be particularly useful.
In glioblastoma, no drug based on a molecular target has been approved. Furthermore, although aberrations in genes such as MGMT and IDH1 may have prognostic significance in glioblastoma (20), these biomarkers have not yet altered clinical practice. However, targets of potential therapeutic relevance are continually being discovered, and tumor genotyping then will likely become very important (21). And many drugs, which are in clinical development or which are approved for other indications, will also be relevant to glioblastoma treatment. Hence, the GLICO model can provide a substrate to screen potential drug targets or therapies by measuring the effects of pharmacologic or genetic interventions on at least tumor cell proliferation, cell death, and invasiveness.
The utility of this model would, however, be increased if it could be brought to sufficient scale in conjunction with advanced robotics. Then both the efficiencies of preclinical drug development and investigations of mechanisms of therapeutic resistance could be accelerated. Realizing these efficiencies would also be a requirement for two additional reasons. First, cancer treatments in combination are more likely to be effective than the same treatments individually. But to have the required numbers of permutations analyzed in a sufficiently rigorous manner to enable robust identification of synergies could easily overwhelm the capacity of the GLICO model system. Second, the enormous costs, long timelines, and low success rates for the clinical development of approved cancer drugs highlight the need for early go/no go decision making in the drug development process. Any in vitro model system that could lead to substantial time and cost savings would attract great interest.
Similarly, scaled up and robotically enhanced versions of the GLICO model could also facilitate its adoption as a useful tool of precision oncology, possibly in combination with next-generation sequencing (21). For clinically meaningful application of the GLICO model in precision oncology, key questions include the ability to obtain matching pairs of samples at primary diagnosis and recurrence, and the time taken to produce a personalized profile of drug testing results so that they are available when needed.
As a concept, biobanks of patient-derived tumor tissues could be used to create populations of ‘in vitro avatars’ for the testing of multiple drugs in co-clinical trials (22). With the necessary technologic refinements, we could envisage several research and clinical scenarios where the GLICO model, or similar approaches, could be applied. For example, if a sufficient number of genetically well-characterized tumor tissues were available, then the resulting large datasets could be used to predict the results for another patient’s tumor, which has similar genetic characteristics but for which the drug testing has not been performed. In different circumstances, it may be possible to carry out a limited number of drug screens quickly enough during the period of administration of first-line therapy so that the screen results could better inform the decision about the next line of an appropriate personalized therapy. In the research setting, other uses could include mass screening exercises, which are designed to discover the antitumor activities in vitro of new chemical entities or repurposed drugs. To maximize the data gained from studies of drug treatment effects, reporter systems may be incorporated into the GLICO model, and opportunities may also exist to use quantitative high-dimensional techniques such as mass cytometry, which may be applied to imaging analysis of microscopic slides or analysis of single cell suspensions, or various omics assays including single-cell RNASeq (23).
Finally, however, the absence of blood vessels and immune cell types that both promote and inhibit antitumor immune responses will limit the utility of the GLICO model for immunotherapy investigations because these investigations require an intact immunocompetent animal. Nevertheless, the fact that the host iPS normal brain organoid is allogeneic to the GSCs may still be useful for the investigation of immunotherapies that are not restricted by MHC type. For example, CAR-T cell therapies comprise genetically engineered T cells that are re-directed toward one or more tumor cell-surface antigens (2). The ability to color these lymphocytes will enable potential quantitative intra-GLICO tracking studies of CAR-T cells (24) as well as studies of interactions between CAR-T cells and tumor targets at the invasive front, which is the origin of recurrent disease.
As our knowledge grows about the tumor microenvironment and our capacity to better mimic this in preclinical in vitro models increases, we anticipate the future discovery of novel therapeutic approaches for glioblastoma treatment based on a better understanding of the interaction of glioblastoma cells with their surrounding microenvironment.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Xian-Xin Qiu [Shanghai Proton and Heavy Ion Center (SPHIC), a.k.a. the Proton and Heavy Ion Center of Fudan University Shanghai Cancer Center (FUSCC), Shanghai, China].
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Linkous A, Balamatsias D, Snuderl M, et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep 2019;26:3203-11.e5. [Crossref] [PubMed]
- Brown MP, Ebert LM, Gargett T. Clinical chimeric antigen receptor-T cell therapy: a new and promising treatment modality for glioblastoma. Clin Transl Immunology 2019;8:e1050. [Crossref] [PubMed]
- Perrin SL, Samuel MS, Koszyca B, et al. Glioblastoma heterogeneity and the tumour microenvironment: implications for preclinical research and development of new treatments. Biochem Soc Trans 2019;47:625-38. [Crossref] [PubMed]
- Zanders ED, Svensson F, Bailey DS. Therapy for glioblastoma: is it working? Drug Discov Today 2019;24:1193-201. [Crossref] [PubMed]
- Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373-9. [Crossref] [PubMed]
- Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015;528:93-8. [Crossref] [PubMed]
- Hubert CG, Rivera M, Spangler LC, et al. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res 2016;76:2465-77. [Crossref] [PubMed]
- da Silva B, Mathew RK, Polson ES, et al. Spontaneous Glioblastoma Spheroid Infiltration of Early-Stage Cerebral Organoids Models Brain Tumor Invasion. SLAS Discov 2018;23:862-8. [Crossref] [PubMed]
- Bian S, Repic M, Guo Z, et al. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 2018;15:631-9. [Crossref] [PubMed]
- Ogawa J, Pao GM, Shokhirev MN, et al. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep 2018;23:1220-9. [Crossref] [PubMed]
- Yi HG, Jeong YH, Kim Y, et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng 2019;3:509-19. [Crossref] [PubMed]
- Oksdath M, Perrin SL, Bardy C, et al. Review: Synthetic scaffolds to control the biochemical, mechanical, and geometrical environment of stem cell-derived brain organoids. APL Bioeng 2018;2:041501. [Crossref] [PubMed]
- Mansour AA, Goncalves JT, Bloyd CW, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 2018;36:432-41. [Crossref] [PubMed]
- Pham MT, Pollock KM, Rose MD, et al. Generation of human vascularized brain organoids. Neuroreport 2018;29:588-93. [Crossref] [PubMed]
- Bergmann S, Lawler SE, Qu Y, et al. Blood-brain-barrier organoids for investigating the permeability of CNS therapeutics. Nat Protoc 2018;13:2827-43. [Crossref] [PubMed]
- Nzou G, Wicks RT, Wicks EE, et al. Human Cortex Spheroid with a Functional Blood Brain Barrier for High-Throughput Neurotoxicity Screening and Disease Modeling. Sci Rep 2018;8:7413. [Crossref] [PubMed]
- Marton RM, Miura Y, Sloan SA, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci 2019;22:484-91. [Crossref] [PubMed]
- Abreu CM, Gama L, Krasemann S, et al. Microglia Increase Inflammatory Responses in iPSC-Derived Human BrainSpheres. Front Microbiol 2018;9:2766. [Crossref] [PubMed]
- Ormel PR, Vieira de Sa R, van Bodegraven EJ, et al. Microglia innately develop within cerebral organoids. Nat Commun 2018;9:4167. [Crossref] [PubMed]
- Szopa W, Burley TA, Kramer-Marek G, et al. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives. Biomed Res Int 2017;2017:8013575. [Crossref] [PubMed]
- Byron SA, Tran NL, Halperin RF, et al. Prospective Feasibility Trial for Genomics-Informed Treatment in Recurrent and Progressive Glioblastoma. Clin Cancer Res 2018;24:295-305. [Crossref] [PubMed]
- Malaney P, Nicosia SV, Dave V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett 2014;344:1-12. [Crossref] [PubMed]
- Eisenstein M. Cellular censuses to guide cancer care. Nature 2019;567:555-7. [Crossref] [PubMed]
- Pageon SV, Govendir MA, Kempe D, et al. Mechanoimmunology: molecular-scale forces govern immune cell functions. Mol Biol Cell 2018;29:1919-26. [Crossref] [PubMed]


