The functions of long non-coding RNAs in colorectal cancer
Introduction
Colorectal carcinoma (CRC) is a common digestive tract tumours. At present, new cases of CRC rank third in malignant tumours, after lung cancer and breast cancer, and mortality ranks fourth among malignant tumours after lung cancer, liver cancer and stomach cancer. Tumour progression and metastasis are the primary causes of death, and many patients have advanced stages with metastasis already having occurred at diagnosis due to the atypical symptoms of CRC (1,2). Hence, chemotherapy, radiotherapy and targeted therapy, even immunotherapy, is required in these patients’ post-surgery, but outcomes remain unsatisfactory. Due to alterations in multiple genes and epigenetics, tumours regulate initiation and inactivation of oncogenes or tumour suppressor genes, enabling tumour cells to acquire stronger invasion abilities, enter the systemic circulation, evade monitoring of immune cells, and finally colonize distant organs. This complex process is necessary for tumour metastasis (3). Therefore, more investigators are aware of the urgent need to further understand the pathogenesis and corresponding molecular mechanisms of CRC, which will contribute to identification of therapeutic targets and diagnostic biomarkers.
Long non-coding RNAs (lncRNAs) are a type of RNA containing more than 200 nucleotides. Much evidence has shown that lncRNAs have the ability to combine with DNA, RNA and proteins. By binding to DNA promoters, lncRNAs can prevent transcription factors from accessing their own promoter binding sites, impeding the transcription of specific genes to regulate epigenetic silencing of target genes. Many studies have also reported that lncRNAs can even serve as molecular scaffolds connecting two or more proteins in functional complexes or to position protein complexes at appropriate cellular compartments. In terms of improving mRNA stability, lncRNAs can target antisense mRNA by directly complementing the sequence, thereby regulating selective splicing processes or protecting the 3'UTR from binding by miRNA. In addition, increasing experiments have confirmed that lncRNAs contain binding sites for miRNAs and represent potential molecular sponges for sequestering the most abundant miRNAs. In other words, lncRNAs may act as competing endogenous RNAs for the function of miRNAs, thus protecting miRNA targets (4). In recent years, lncRNAs have been increasingly found to play critical roles in a wide range of biological processes, such as cancer cell proliferation, invasion, apoptosis, metastasis and chemoresistance, including in CRC. Therefore, lncRNAs have been identified as future diagnostic, therapeutic, and/or prognostic cancer biomarker (5-7). The objective of this review was to summarize lncRNAs related to the Wnt/β-catenin pathway, epithelial mesenchymal transition, epigenetic regulation, angiopoiesis, and chemoresistance.
LncRNAs related to the Wnt/β-catenin pathway
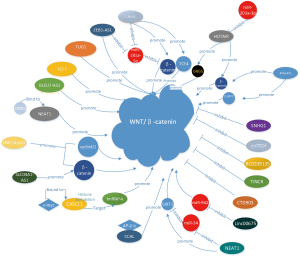
Wnt/β-catenin signalling is critical for CRC initiation (8). Casein kinase 1 (CK1), glycogen synthase kinase 3β (GSK-3β), Axin, protein phosphatase 2A (PP2A), and APC and activator protein 2α (AP-2α) are all significant in the Wnt signalling pathway (9). Dysfunction of these factors leads to activation of Wnt signalling by converting free β-catenin accumulated in the nucleus and then stimulating the E2F transcription factor-4 (E2F4), the proto-oncogene c-Myc, and cyclin D1 (CCND1), which are targets of the lymphoid enhancing factor (LEF)/T-cell factor (TCF) (10).
The lncRNA colorectal neoplasia differentially expressed (CRNDE), which is transcribed from chromosome 16 on the strand opposite the adjacent IRX5 gene, is a novel biomarker in CRC and other solid cancers. CRNDE has been shown to be elevated in CRC cells and promotes the proliferation of CRC cells via miR-181a-5p-mediated regulation of Wnt/β-catenin signalling. Han et al. (11) reported that β-catenin and TCF4 are inhibitory targets of miR-181a-5p, and Wnt/β-catenin signalling is activated by both CRNDE overexpression and miR-181a-5p knockdown. Finally, they indicated that CRNDE may represent a prominent therapeutic target in CRC.
Zhang et al. (12) reported that cancer susceptibility candidate 11 (CASC11) is upregulated in CRC tissues, further demonstrating that c-Myc directly binds to the promoter regions of CASC11 and enhances CASC11 expression by increasing promoter histone acetylation, and finally, CASC11 aligns with heterogeneous ribonucleoprotein K (hnRNP-K) to activate Wnt/β-catenin signalling in CRC cells. Increasing expression of CASC11 conclusively leads to proliferation and metastasis of CRC.
Another oncogene, the lncRNASLCO4A1-AS1, which has been confirmed to have high expression in various cancers, especially in CRC, activates Wnt signalling to initiate tumorigenesis. Yu et al. (13) discovered that expression of the lncRNASLCO4A1-AS1 is significantly upregulated in CRC tissue and cell lines, which activates Wnt/β-catenin signalling via enhancing the stability of β-catenin. Finally, accumulation of β-catenin protein in the nucleus enhances proliferation, migration and invasion in SLCO4A1-AS1-depleted CRC cells, suggesting that SLCO4A1-AS1 may represent a novel therapeutic target.
The lncRNA NEAT1 (nuclear paraspeckle assembly transcript 1), which is located at 11q13.1, is upregulated in CRC cells. NEAT1 was identified as a novel regulator of the Wnt/β-catenin pathway in CRC. Many studies have uncovered that NEAT1 may act as a competing endogenous RNA (ceRNA) via sequestering miRNAs (14). In Yang Luo’s research, they deduced that NEAT1 was abnormally highly expressed and significantly activated the Wnt/β-catenin signalling pathway. It primarily functions as a ceRNA for miR-34 to repress SIRT1 expression. Taken together, their findings demonstrate that the lncRNA NEAT1 may represent a prognostic biomarker and a potential therapeutic target in CRC (15).
In contrast, many other lncRNAs, such as LINC00675, CTD903, SNHG1, TINCR and BC0209135 (16-20), were recently proven to be tumour suppressors in CRC. High levels of these factors were suggested to inhibit Wnt/β-catenin signalling in CRC (Figure 1).
LncRNAs affect epithelial mesenchymal transition (EMT)
EMT is an important way to promote tumour cell metastasis. Its pathological features include epithelial cells losing their polarity and losing adhesion to other cells, while gaining the ability to migrate and infiltrate, making quiescent epithelial cells transform into motile mesenchymal cells (21,22). Characteristic changes of EMT include downregulation of E-cadherin and upregulation of N-cadherin, ZEB1, ZEB2, snail1 and snail2. Accumulating evidence has confirmed that EMT plays a vital role in CRC metastasis, and many lncRNAs induce CRC metastasis via EMT (Figure 2). These factors can be divided into two categories, one of which is upregulated in CRC and is defined as oncogene, while another classic lncRNA is downregulated and is defined as a tumour suppressor gene, affecting EMT and inducing metastasis.
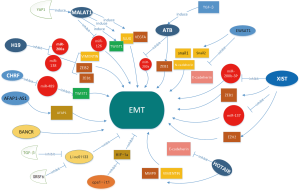
As the first imprinted gene to be discovered, the lncRNA H19 gene is mapped to chromosome 11 in humans and was defined as a maternally expressed imprinted gene that plays an important role in mammalian development (23). Ding et al. (24) reported that lncRNA H19 is upregulated in CRC compared to adjacent normal colorectal mucosa cells and that expression of E-cadherin is increased, while expression of vimentin and snail is decreased. Liang et al. (25) found that H19 expression was significantly increased in mesenchymal-like cancer cells and CRC tissues, significantly stimulating EMT progression and inducing tumour growth both in vivo and in vitro. Furthermore, they confirmed that H19 adsorbs miR-138 and miR-200a by sponge-like adsorption, inhibiting their function and resulting in unrestricted production of their endogenous targets, Vimentin, ZEB1 and ZEB2, which are core marker genes of mesenchymal cells. Taken together, these experimental phenomena suggest that lncRNA H19 regulates expression of multiple EMT-related genes by acting as a competitive endogenous RNA, potentially constituting a link between regulation of miRNA networks and EMT progression. In addition, Liang et al. (26) found that the sTLR4/MD-2 complex inhibits CRC migration and invasion by lncRNA H19 downregulation both in vitro and in vivo.
LncRNA EWSAT1 (Ewing sarcoma-associated transcript 1), which is located at chromosome 15, has been confirmed to be highly expressed in various tumour tissues, promoting the occurrence and development of cancers, including CRC. EWSAT1 is overexpressed in CRC tissues compared to matched adjacent normal tissues, and higher expression of EWSAT1 promotes EMT by reducing E-cadherin expression and increasing snail1, snail2 and N-cadherin expression. Therefore, EWSAT1 was deduced to be an oncogene that promotes metastasis via initiation of EMT progression (27).
MALAT1 (metastasis associated lung adenocarcinoma transcript 1), a transcriptional regulator for numerous genes, including genes involved in cancer metastasis and invasion, is primarily localized in the cytoplasm of colon cancer cells according to cell fractionation assays. The lncRNA MALAT-1 is reportedly upregulated by YAP1 in CRC tissues, stimulating expression of metastasis-associated molecules, such as TWIST, SLUG, and VEGFA, by sponging miR-126-5p in CRC to affect EMT (28).
LncRNA-ATB is located on human chromosome 14:19,858,667–19,941,024. lncRNA-ATB has been indicated to be a transcript with two exons spanning more than 80 kb. lncRNA-ATB is upregulated in response to overexpression of transforming growth factor-β (TGF-β), and many studies have shown that lncRNA-ATB induces EMT, leading to the promotion of tumour progression, i.e., enhancement of proliferation, migration, and invasion (29). Yue et al. (30) demonstrated that expression of lncRNA ATB is significantly upregulated in CRC tissue and cell lines compared to adjacent normal mucosa; however, E-cadherin was decreased in cancer tissues. Reduced levels of E-cadherin result in decreased adhesion strength between cells in a tissue, leading to increased cellular activity and facilitating cancer cell invasion to surrounding tissues through the basement membrane and into systemic circulation. Further studies (31) showed that lncRNA ATB induces ZEB1 expression by inhibiting miR-200s in tumour cells to affect EMT in colorectal neoplasm cells.
Many studies have reported that lncRNA X-inactive specific transcript (lncRNA XIST) is highly expressed in multiple tumours. Chen et al. (32) reported that expression of lncRNA XIST is upregulated in both CRC cell lines and tissues. High expression of lncRNA XIST promotes CRC cell proliferation and invasion through EMT progression, and further study confirmed that lncRNA XIST directly binds to miR-200b-3p, increasing expression of ZEB1, a direct target of miR-200b-3p. Liu et al. (33) reported that lncRNA XIST promotes EMT progression by regulating the miR-137-EZH2 axis.
Another lncRNA, HOTAIR (HOX transcript antisense RNA), which is located on chromosome 12q13.13, has been considered a proto-oncogene in a variety of tumours and was found to demonstrate trans-transcriptional regulatory function. To further investigate its biological functions, studies have shown that lncRNA HOTAIR is elevated in CRC tissues and promotes EMT progression by increasing expression of vimentin and matrix metallopeptidase9and decreasing expression of E-cadherin. Therefore, HOTAIR may serve as a very promising predictor and therapeutic target for CRC (34).
As a well-known lncRNA, actin filament associated protein 1 antisense RNA1 (AFAP1-AS1) is an antisense RNA gene encoding AFAP1, which is also highly expressed in CRC. Western blot results showed that expression of E‐cadherin was elevated, however, expression of N‐cadherin, vimentin, and fibronectin was reduced when AFAP1‐AS1 was knocked-down, confirming that AFAP1-AS1 participates in cell proliferation, colony formation, migration and invasion via the EMT pathway (35).
The BRAF-activated non-coding RNA (BANCR) is a lncRNA with a length of 693 bp located on chromosome 9, and numerous studies have shown that BANCR is involved in many biological processes, including EMT progression. It has been proven that BANCR is upregulated in CRC and induces EMT via a mitogen-activated protein kinase kinase/extracellular signal-regulated kinase-dependent mechanism, enhancing G0/G1 cell cycle arrest and apoptosis by regulating p21. These findings indicate that BANCR plays an important role in CRC and may represent a novel and useful biomarker (6,36).
The lncRNAs expounded above are all upregulated as oncogenes that promote EMT; however, there are many lncRNAs that are downregulated in CRC and are defined as tumour suppressor genes that inhibit EMT progression.
LINC01133, another novel lncRNA with reduced expression in CRC, was shown by Kong et al. (37) to be downregulated by TGF-β, which inhibited EMT and metastasis in CRC cells. Furthermore, they found that EMT was affected by LINC01133 in CRC cells and was dependent on the existence of SRSF6. As a target mimic, Linc01133 was defined as a prognostic biomarker and an effective target for anti-metastatic therapies for CRC.
LncRNA CPS1 intronic transcript 1 (lncRNA CPS1-IT1), was recently identified as a tumour suppressor, and Zhang et al. (38) found that lncRNACPS1-IT1is significantly decreased in CRC, resulting in worse prognosis. As a tumour suppressor, CPS1 reverses EMT of tumours by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α in CRC. These findings suggest that lncRNA CPS1-IT1 affects EMT progression and inhibits tumour invasion and metastasis.
Epigenetic regulation in CRC
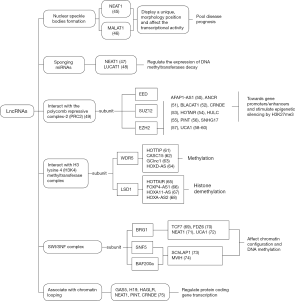
Recent genomic analyses have uncovered epigenetic regulation as a major driver of tumorigenesis (39), lncRNAs can be cytoplasmic and/or nuclear, and nuclear lncRNAs reportedly regulate histone and DNA modifications (40). Therefore, at both the histone and DNA methylation levels, many lncRNAs have been found to bind to epigenetic complexes and play roles in controlling the DNA modification system in CRC cells (41-44) (Figure 3).
The nucleoli and paraspeckles maintain a unique morphology in the nucleus and play vital roles in transcriptional activity. NEAT1 and MALAT1, which are overexpressed in CRC tissues, have been shown to play a significant role in the formation and organization of nuclear speckle bodies, leading to poor disease prognosis (45,46). Another lncRNAs, such as NEAT1 and LUCAT1, interfere with miRNA and DNMT, affecting DNA methylation (47,48).
LncRNAs also may interact with multicomb inhibitory complex 2 (PRC 2), in which the three main subunits are embryonic ectoderm development (EED), SUZ12, and ZEST homology enhancer (EZH2) in embryonic ectoderm development. Approximately 20–30% of lncRNA genes have been confirmed by RNA immunoprecipitation to interact with PRC2 by binding to SUZ12 and EZH2 (49). In CRC cells, there are many lncRNAs, such as AFAP1-AS1 (50), ANCR (51), BLACAT (52), CRNDE (53), HOTAIR (54), HULC (55), PINT (56), SNHG17 (57), UCA1 (58-60), that bind to EZH2. LncRNAs solicit PRC2 from gene promoters/enhancers, which are genetically silenced by trimethyl lysine 27 (K27me3) stimulation of histone H3 (H3K27me3). In addition to the lncRNA/EZH2 combination, there are some lncRNAs, such as HOTTIP (61), CASC15 (62), GClnc1 (63), and HOXD-AS (64), that interact with the adaptor protein WDR5 from the histone H3 lysine4 (H3K4) methyl transferase complex. In contrast, HOTTAIR (65), FOXP4-AS1 (66), HOXA11-AS (67), and HOXA-AS2 (68) interact with histone lysine-specific demethylase 1 (LSD1), which may also affect histone demethylation.
LncRNAs interact with switching defective sucrose/non-fermenting (SWI/SNF) complexes, affecting chromatin configuration and DNA methylation. SWI/SNF complexes have three subunits, Brahma related gene 1 (BRG1), SNF5 and BAF200a. It has been shown that lncTCF7 (69), lncFDZ6 (70), NEAT1 (71), and UCA1 (72) bind to BRG1 and SChLAP1 (73), while MVIH (74) binds to SNF5 and BAF200a.
Antisense coding lncRNAs, such as HAGLR, GAS5, NEAT1, H19, PINT, and CRNDE, which were reportedly associated with chromatin looping, directly suppress the transcription of sense coding genes, and their overexpression leads to poor prognosis in CRC (75).
Recently, many studies have confirmed specific functions of a number of p53-regulated lncRNAs, including LincRNA-p21, PURPL, PANDA, NEAT1, DINO, PINT, LED, PR-lncRNA-1, Linc-475 and PINCR. Li et al. (76) investigated PURPL, the lncRNA that is regulated by p53, which inhibits basal p53 levels and facilitates tumorigenicity in CRC via associating with MYBBP1A and inhibiting formation of the p53-MYBBP1A complex.
Another lncRNA, colon cancer-associated transcript 2 (CCAT2), is an RNA with the length of 1,752 bp located on chromosome 8q24.21. This genomic locus harbors the SNP rs6983267 (77), which plays a vital role in colorectal cancer. It has been revealed that lncRNA CCAT2 expression levels remarkedly elevated in CRC tissues compared to paired normal mucosae. In Ling et al.’s experiment (78), they confirmed that high expression of CCAT2 is one of the primary characteristics of microsatellite-stable (MSS) CRCs that leads to chromosomal instability (CIN), and the centrosome defects they observed may lead to abnormal chromosome breakage and fusion, eventually leading to aneuploidy. Further studies found that overexpression of CCAT2 leads to higher expression of the MYC gene, as well as expression of downstream MYC protein-coding gene targets, such as BAX, CDC25A and CDKN2A, as well as microRNA targets, such as MIR17HG.Taken together, CCAT2 is a useful diagnostic indicator and therapeutic target.
Accumulating evidence has shown that expression levels of lncRNAs are positively correlated with patient prognosis in CRC, indicating that lncRNAs play a critical role in modulating the cancer epigenome and may be significant targets for CRC diagnosis and therapy.
LncRNAs and angiogenesis
Ample evidence indicates that tumour growth and metastasis are accompanied by the formation of tumour blood vessels. Growth of tumour blood vessels not only provides nutrition for tumours but also provides a pathway for metastasis (79). There is increasing evidence that lncRNAs play a vital role in tumour angiogenesis (80). However, in CRC, there are no studies examining the effect of lncRNA on angiogenesis. Using enzyme-linked immunosorbent assay (ELISA) and RNA pull-down (RIP) methods, lncRNA MVIH was shown to attenuate angiogenesis via inhibiting the secretion of PGK1 (phosphoglycerate kinase 1), a key factor for angiogenesis (81). The lncRNA MALAT1 has also been shown to cause tumour angiogenesis. MALAT1 activates angiogenesis by regulating vascular density and vasodilation (82,83), while MEG3 is reported to reduce the formation of tumour blood vessels by affecting expression of VEGF (84). Recently, H19, LincRNA-p21, TUG1 and HOTAIR have all been confirmed to play important roles in tumour angiogenesis. Interestingly, these lncRNAs have also been shown to play important roles in the development of CRC. Therefore, these lncRNAs are likely to be involved in the formation of blood vessels in colorectal tumours, which is a hot topic for further research.
Chemoresistance
Chemotherapy resistance is an important reason for the failure of advanced CRC treatment. In recent years, many studies have shown that lncRNAs play an important role in tumour chemotherapy resistance. Li et al. (85) confirmed that lncMEG3 is downregulated in oxaliplatin-resistant CRC patients and has considerable potential for identification of responsive and non-responsive patients. In addition, in patients with CRC treated with oxaliplatin, those with reduced serum MEG3 expression experience increased chemoresistance and reduced survival. They demonstrated that expression of lncRNA MEG3 is reduced in CRC, with consequent poor response to therapy. The main mechanism for this effect involves lncRNA MEG3 increasing chemosensitivity by enhancing oxaliplatin-induced apoptosis. Therefore, overexpression of MEG3 may be a novel therapeutic direction for CRC patients to reverse oxaliplatin resistance.
Another lncRNA, UCA1, weakens the response to 5-fluorouracil (5-FU) chemosensitivity in CRC by reducing apoptosis via inhibiting miR-204-5p. The UCA1-miR-204-5p-CREB1/BCL2/RAB22A regulatory network plays a vital role in pathogenesis and chemoresistance in CRC patients (86). In addition, Wu et al. (87) posited that UCA1 sequesters miR-204 by regulating HMGA2 in CRC cells, affecting chemoresistance. Lee et al. (88) showed that upregulation of the lncRNA snaR increases cell death after treatment with 5-FU, which manifests as snaR loss increasing resistance to 5-FU in CRC. Linc00152 sequesters miR-193a-3p and elevates levels of ERBB4, which contribute to oxaliplatin chemosensitivity in colon cancer (89). Li et al. verified that lncRNA TUG1 is significantly increased in CRC, causing methotrexate resistance. They also found that lncRNA TUG1 sequesters miR-186, and cytoplasmic polyadenylation element binding protein 2 (CPEB2) is negatively correlated with expression levels of miR-186. As such, they propose that lncRNA TUG1 mediates MTX resistance in CRC via the miR-186/CPEB2 axis. As previously mentioned, the lncRNA CCAL induces CRC cell multidrug-resistance through activation of Wnt/β-catenin signalling by inhibiting AP-2α and leading to upregulation of MDR1/P-gp expression (90). AP-2α serves as an oncosuppressive protein by interacting with APC and β-catenin.
Conclusions
Accumulating evidence has proven that lncRNAs play significant roles in tumorigenesis and development of CRC. In this review, we summarized abnormal expression of lncRNAs that heavily influence different biological progresses, including the Wnt/β-catenin signalling pathway, EMT, angiogenesis, epigenetic regulation and chemoresistance. In CRC diagnosis, due to their convenience and non-invasiveness, biomarkers in plasma and serum perform a vital function for comparing colonoscopy examinations. However, in the current blood test, the best biomarkers, carcinoembryonic antigen (CEA) and carbohydrate antigen19-9 (CA19-9), exhibit low specificity and sensitivity, particularly in early stage CRC. Therefore, the main priority is identifying novel biomarkers to reliably detect early CRC and relapse in patients’ post-surgery. First, lncRNAs are regarded as targets of tumour prediction. Abnormal expression of lncRNAs affects tumour development. Intriguingly, recent studies have shown that some lncRNAs can be detected in the serum of cancer patients. For instance, detection of lncRNA CRNDE-h in exosomes shed light on utilizing exosomal CRNDE-h as a non-invasive serum-based tumour marker for diagnosis and prognosis of CRC (91). Several other dysregulated lncRNAs, such as BANCR, NR_029373, NR_026817, and NR_034119, are present in both cancer tissues and patient serum samples (92). There is concern that a single lncRNA may not be sufficient for predicting CRC prognosis. However, if several lncRNA combinations that were known to be involved in CRC progression were identified, it might solve the issue. It is also worth considering whether there are different sub-classifications of lncRNA expression in patients with cancer at different sites in CRC. Barbagallo et al. recently found that TUG1, a tumour suppressor gene, is decreased in CRC tissues but increased in exosomes, while UCA1, which acts as an oncogene, displays the opposite behaviour. This observation suggests that tumour cells secrete lncRNAs through exosomes to protect themselves from TUG1 tumour-suppressive activity. The oncogenic function of UCA1 is critical to tumour progression, inducing tumour cell proliferation by limiting secretion of UCA1.It is well known that use of a signature of multiple biomarkers is preferable, as it would increase diagnostic and prognostic accuracy of a single test. Barbagallo et al. calculated a combination ROC curve considering two lncRNAs with opposite trends in expression that may increase the sensitivity and specificity compared to a single biomarker (93,94). As found in gene mutations before, such as NRAS, BRAS and KRAS genes that well known to be vital in the advancement and development of CRC, increasing lncRNAs are being discovered, and they have been shown to have distinct functions in tumorigenesis and development, such as influencing epigenetics, gene transcription, chemical resistance and so on, including in CRC research. In conclusion, lncRNAs play crucial roles as predictors of cancer. Additional studies with large cohort of CRC patients should be performed in the future to expand these findings.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.23). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [Crossref] [PubMed]
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62:220-41. [Crossref] [PubMed]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011;331:1559-64. [Crossref] [PubMed]
- Ragusa M, Barbagallo C, Brex D, et al. Molecular Crosstalking among Noncoding RNAs: A New Network Layer of Genome Regulation in Cancer. Int J Genomics 2017;2017:4723193. [Crossref] [PubMed]
- Yin DD, Liu ZJ, Zhang E, Kong R, et al. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol 2015;36:4851-9. [Crossref] [PubMed]
- Shi Y, Liu Y, Wang J, et al. Downregulated long noncoding RNA BANCR promotes the proliferation of colorectal cancer cells via downregualtion of p21 expression. PLoS One 2015;10:e0122679. [Crossref] [PubMed]
- Lian Y, Wang J, Feng J, et al. Long non-coding RNA IRAIN suppresses apoptosis and promotes proliferation by binding to LSD1 and EZH2 in pancreatic cancer. Tumour Biol 2016;37:14929-37. [Crossref] [PubMed]
- Wilkins JA, Sansom OJ. C-Myc is a critical mediator of the phenotypes of Apc loss in the intestine. Cancer research 2008;68:4963-6. [Crossref] [PubMed]
- Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene 2006;25:7482-91. [Crossref] [PubMed]
- Sansom OJ, Reed KR, Hayes AJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 2004;18:1385-90. [Crossref] [PubMed]
- Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017;16:9. [Crossref] [PubMed]
- Zhang Z, Zhou C, Chang Y, et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett 2016;376:62-73. [Crossref] [PubMed]
- Yu J, Han Z, Sun Z, et al. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J Exp Clin Cancer Res 2018;37:222. [Crossref] [PubMed]
- Guo Y, Zhang H, Xie D, et al. Non-coding RNA NEAT1/miR-214-3p contribute to doxorubicin resistance of urothelial bladder cancer preliminary through the Wnt/β-catenin pathway. Cancer Manag Res 2018;10:4371-80. [Crossref] [PubMed]
- Luo Y, Chen JJ, Lv Q, et al. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/β-catenin signaling pathway. Cancer Lett 2019;440-441:11-22. [Crossref] [PubMed]
- Ma S, Deng X, Yang Y, et al. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/β-catenin signaling in cervical cancer. Biomed Pharmacother 2018;108:1686-93. [Crossref] [PubMed]
- Yuan Z, Yu X, Ni B, et al. Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/β-catenin signaling and predicts favorable prognosis. Int J Oncol 2016;48:2675-85. [Crossref] [PubMed]
- Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem Biophys Res Commun 2018;495:238-45. [Crossref] [PubMed]
- Zheng Q, Lin Y, Chen P, et al. The long noncoding RNA BC0209135 inhibits the cell invasion through Wnt/β-catenin signaling in colorectal cancer. Eur Rev Med Pharmacol Sci 2018;22:3763-70. [PubMed]
- Zhang ZY, Lu YX, Zhang ZY, et al. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget 2016;7:22639-49. [PubMed]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131-42. [Crossref] [PubMed]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008;14:818-29. [Crossref] [PubMed]
- Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature cell biology 2012;14:659-65. [Crossref] [PubMed]
- Ding D, Li C, Zhao T, et al. LncRNA H19/miR-29b-3p/PGRN Axis Promoted Epithelial-Mesenchymal Transition of Colorectal Cancer Cells by Acting on Wnt Signaling. Mol Cells 2018;41:423-35. [PubMed]
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015;6:22513-25. [Crossref] [PubMed]
- Liang W, Zou Y, Qin F, et al. sTLR4/MD-2 complex inhibits colorectal cancer migration and invasiveness in vitro and in vivo by lncRNA H19 down-regulation. Acta Biochim Biophys Sin (Shanghai) 2017;49:1035-41. [Crossref] [PubMed]
- Zhang R, Li JB, Yan XF, et al. Increased EWSAT1 expression promotes cell proliferation, invasion and epithelial-mesenchymal transition in colorectal cancer. Eur Rev Med Pharmacol Sci 2018;22:6801-8. [PubMed]
- Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 2019;38:2627-44. [Crossref] [PubMed]
- Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov 2012;11:790-811. [Crossref] [PubMed]
- Yue B, Qiu S, Zhao S, et al. LncRNAATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol 2016;31:595-603. [Crossref] [PubMed]
- Iguchi T, Uchi R, Nambara S, et al. A Long Noncoding RNA, lncRNA-ATB, Is Involved in the Progression and Prognosis of Colorectal Cancer. Anticancer Res 2015;35:1385-8. [PubMed]
- Chen DL, Chen LZ, Lu YX, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis 2017;8:e3011. [Crossref] [PubMed]
- Liu X, Cui L, Hua D. Long non-coding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol Res 2018;27:99-106. [Crossref] [PubMed]
- Wu ZH, Wang XL, Tang HM, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep 2014;32:395-402. [Crossref] [PubMed]
- Wang F, Ni H, Sun F, et al. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother 2016;81:152-9. [Crossref] [PubMed]
- Guo Q, Zhao Y, Chen J, et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelialmesenchymal transition. Oncol Lett 2014;8:869-875. [Crossref] [PubMed]
- Kong J, Sun W, Li C, et al. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett 2016;380:476-84. [Crossref] [PubMed]
- Zhang W, Yuan W, Song J, et al. LncRna CPS1-IT1 Suppresses Cell Proliferation, Invasion and Metastasis in Colorectal Cancer. Cell Physiol Biochem 2017;44:567-80. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Beckedorff FC, Amaral MS, Deocesanopereira C, et al. Long non-coding RNAs and their implications in cancer epigenetics. Biosci Rep 2013; [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014;54:777-90. [Crossref] [PubMed]
- Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci 2017;108:1927-33. [Crossref] [PubMed]
- Chaudhary R, Gryder B, Woods WS, et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife 2017; [Crossref] [PubMed]
- Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33:717-26. [Crossref] [PubMed]
- Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol 2013;10:456-61. [Crossref] [PubMed]
- Li Y, Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am J Cancer Res 2018;8:81-90. [PubMed]
- Yoon JH, You BH, Park CH, et al. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett 2018;417:47-57. [Crossref] [PubMed]
- Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667-72. [Crossref] [PubMed]
- Tang J, Zhong G, Wu J, et al. Long noncoding RNA AFAP1-AS1 facilitates tumor growth through enhancer of zeste homolog 2 in colorectal cancer. Am J Cancer Res 2018;8:892-902. [PubMed]
- Yang ZY, Yang F, Zhang YL, et al. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark 2017;18:95-104. [Crossref] [PubMed]
- Su J, Zhang E, Han L, et al. Long non-coding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis 2017;8:e2665. [Crossref] [PubMed]
- Ding J, Li J, Wang H, et al. Long non-coding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis 2017;8:e2997. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Yang XJ, Huang CQ, Peng CW, et al. Long noncoding RNA HULC promotes colorectal carcinoma progression through epigenetically repressing NKD2 expression. Gene 2016;592:172-8. [Crossref] [PubMed]
- Marín-Béjar O, Marchese FP, Athie A, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 2013;14:R104. [Crossref] [PubMed]
- Ma Z, Gu S, Song M, et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst 2017;13:2350-61. [Crossref] [PubMed]
- Cai Q, Jin L, Wang S, et al. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget 2017;8:47957-68. [Crossref] [PubMed]
- Wang ZQ, Cai Q, Hu L, et al. Long non-coding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis 2017;8:e2839. [Crossref] [PubMed]
- Hu JJ, Song W, Zhang SD, et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep 2016;6:23521. [Crossref] [PubMed]
- Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120-4. [Crossref] [PubMed]
- Wu Q, Xiang S, Ma J, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol 2018;12:799-813. [Crossref] [PubMed]
- Sun TT, He J, Liang Q, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov 2016;6:784-801. [Crossref] [PubMed]
- Gu P, Chen X, Xie R, et al. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol Ther 2017;25:1959-73. [Crossref] [PubMed]
- Xia M, Yao L, Zhang Q, et al. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget 2017;8:19795-802. [PubMed]
- Yang L, Ge D, Chen X, et al. FOXP4-AS1 participates in the development and progression of osteosarcoma by downregulating LATS1 via binding to LSD1 and EZH2. Biochem. Biophys. Res Commun 2018;502:493-500. [Crossref] [PubMed]
- Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res 2016;76:6299-310. [Crossref] [PubMed]
- Ding J, Xie M, Lian Y, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis 2017;6:e288. [Crossref] [PubMed]
- Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015;16:413-25. [Crossref] [PubMed]
- Chen Z, Gao Y, Yao L, et al. LncFZD6 initiates Wnt/beta-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene 2018;37:3098-112. [Crossref] [PubMed]
- Kawaguchi T, Hirose T. Chromatin remodeling complexes in the assembly of long non-coding RNA-dependent nuclear bodies. Nucleus 2015;6:462-7. [Crossref] [PubMed]
- Wang X, Gong Y, Jin BO, et al. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol Rep 2014;32:1281-90. [Crossref] [PubMed]
- Prensner JR, Iyer MK, Sahu A, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 2013;45:1392-8. [Crossref] [PubMed]
- Cheng S, Wang L, Deng CH, et al. ARID1A represses hepatocellular carcinoma cell proliferation and migration through lncRNA MVIH. Biochem Biophys Res Commun 2017;491:178-82. [Crossref] [PubMed]
- Amaral PP, Leonardi T, Han N, et al. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol 2018;19:32. [Crossref] [PubMed]
- Li XL, Subramanian M, Jones MF, et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep 2017;20:2408-23. [Crossref] [PubMed]
- Haiman CA, Le Marchand L, Yamamato J, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet 2007;39:954-6. [Crossref] [PubMed]
- Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013;23:1446-61. [Crossref] [PubMed]
- Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol 1992;3:65-71. [PubMed]
- Khorshidi A, Dhaliwal P, Yang BB. Noncoding RNAs in Tumor Angiogenesis. Adv Exp Med Biol 2016;927:217-41. [Crossref] [PubMed]
- Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012;56:2231-41. [Crossref] [PubMed]
- Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014;114:1389-97. [Crossref] [PubMed]
- Thum T, Fiedler J. LINCing MALAT1 and angiogenesis. Circ Res 2014;114:1366-8. [Crossref] [PubMed]
- Su W, Xie W, Shang Q, et al. The Long Noncoding RNA MEG3 Is Downregulated and Inversely Associated with VEGF Levels in Osteoarthritis. Biomed Res Int 2015;2015:356893. [Crossref] [PubMed]
- Li L, Shang J, Zhang Y, et al. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol Rep 2017;38:1383-92. [Crossref] [PubMed]
- Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep 2016;6:23892. [Crossref] [PubMed]
- Wu H, Liang Y, Shen L. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol Open 2016;5:563-70. [Crossref] [PubMed]
- Lee H, Kim C, Ku JL, et al. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol Cells 2014;37:540-6. [Crossref] [PubMed]
- Yue B, Cai D, Liu C, et al. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther 2016;24:2064-77. [Crossref] [PubMed]
- Li Q, Lohr CV, Dashwood RH. Activator protein 2alpha suppresses intestinal tumorigenesis in the Apc(min) mouse. Cancer Lett 2009;283:36-42. [Crossref] [PubMed]
- Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectalcancer. Oncotarget 2016;7:85551-63. [Crossref] [PubMed]
- Wang R, Du L, Yang X. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol 2016;142:2291-301. [Crossref] [PubMed]
- Borrebaeck CA. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat Rev Cancer 2017;17:199-204. [Crossref] [PubMed]
- Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol Ther Nucleic Acids 2018;12:229-41. [Crossref] [PubMed]