SE translation mRNA overexpression: a potential prognostic predictor in breast cancer
Introduction
Breast cancer is one of the commonest tumors accounting for about 1 in 4 cancer cases among women (1). This disease is well known by its high morbidity and mortality. It was reported that the number of diagnosed patients ranked the top in all types of female tumors (1), among which one of six women died of this disease (2). Therefore, breast cancer has always been the focus issue for scientific researchers. In our previous studies, SE translation (SET) was found to be overexpressed both in mRNA and protein levels in paclitaxel-resistant breast cancer cells, implying that SET might be a potential biomarker of drug resistance (3).
SET gene was located on human chromosome 9, the protein of which was commonly expressed in various nucleuses of tissues (4) and took part in multiple vital physiological and pathological processes, such as cell cycle regulation, cell migration and metastasis, gene transcription, DNA repair, etc. (5). Numerous studies have demonstrated that abnormal expression of SET was related to different kinds of diseases like Alzheimer (6), polycystic ovarian syndrome and hormone-related tumors. SET overexpression was associated to tumor progression and metastasis, whereas its low expression could be linked to the inhibition of tumor cell proliferation (7,8). Moreover, SET was associated with poor prognosis and Oxaliplatin resistance in metastatic colorectal cancer patients which indicates that SET may act as a potential oncogene (9). SET plays such an important role in cancer progression. However, whether SET can lead to poor prognosis in breast cancer is lack of recognition. In this study, we investigated the association among SET mRNA, clinicopathological parameters, and prognosis of breast cancer patients in a large cohort. In the meantime, we also explored the functions of SET and its related genes in the disease process.
Methods
Patients and materials
All the clinical data of breast cancer patients were obtained from molecular taxonomy of breast cancer international consortium (METABRIC) database, including SET mRNA expression, diagnosed age, tumor size, neoplasm histologic grade, tumor stage, ER status, PR status, HER-2 status, inferred menopausal state and overall survival (OS). Besides, the expression network of SET and its related genes were searched from cBioPortal website (http://www.cbioportal.org/) for the exploration of their function in the progression of breast cancer.
Cut-off score of SET mRNA expression
The cut-off score of SET mRNA expression was calculated by X-tile 3.6.1 software (Yale University, New Haven, CT, USA). We input OS and SET mRNA expression scores of breast cancer patients which were acquired from METABRIC database, then set the latter index as the marker. Finally we got the cut-off score, which divided mRNA expression into high and low expression.
Statistical analysis
Chi-square test was used to analyze the relationship between different clinicopathological characteristics and SET mRNA expression for breast cancer patients. This process was followed by multivariate logistic regression in order to figure out independent influence factors for SET mRNA expression. COX regression model was then applied to investigate the influence of relative factors for OS. Survival curves were generated using the Kaplan-Meier method to examine the prognostic value of SET. A two-tailed P value of <0.05 was considered statistically significant. All data were analyzed using SPSS statistics for Windows version 20.0 (SPSS Inc., Chicago, IL, USA).
Results
Data acquirement and classification
Among 2,509 breast cancer samples in the database, we selected 1,904 cases with complete information for analysis. After calculated by X-tile software, the cutoff score of SET mRNA was −0.05, which was used to divide breast cancer patients into high expression group (score ≥−0.05) and low expression group (score <−0.05).
Relationship between SET mRNA expression and clinicopathological characteristics
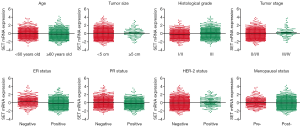
As Figure 1 shows, we divided each clinicopathological characteristic into two types. Chi-square test was then conducted to assess the correlation between clinicopathological factors and SET mRNA expression level. The results are listed in Table 1. The expression level of SET mRNA was significantly related to diagnosed age (P=0.010), tumor size (P=0.021), neoplasm histologic grade (P<0.001), tumor stage (P=0.005), ER status (P<0.001), PR status (P<0.001), HER-2 status (P=0.044), and inferred menopausal status (P=0.041). Multivariate logistic regression indicated that tumor histological grade (P<0.001) and ER status (P<0.001) were independent influence factors of SET mRNA expression level for breast cancer patients (Table 2). The cases with poor tumor tissue differentiation or negative ER tended to overexpress SET mRNA.
Table 1
| Characteristics | Cases | mRNA expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Age | ||||
| <60 | 842 | 395 | 447 | 0.010* |
| ≥60 | 1,062 | 561 | 501 | |
| Tumor size (cm) | ||||
| <5 | 1,744 | 888 | 856 | 0.021* |
| ≥5 | 142 | 58 | 84 | |
| Histological grade | ||||
| I/II | 905 | 509 | 396 | <0.001*** |
| III | 927 | 403 | 524 | |
| Tumor stage | ||||
| 0/I/II | 1,279 | 674 | 605 | 0.005** |
| III/IV | 124 | 49 | 75 | |
| ER status | ||||
| Negative | 445 | 152 | 293 | <0.001* |
| Positive | 1,459 | 804 | 655 | |
| PR status | ||||
| Negative | 895 | 400 | 495 | <0. 001*** |
| Positive | 1,009 | 556 | 453 | |
| Her-2 status | ||||
| Negative | 1,668 | 852 | 816 | 0.044* |
| Positive | 236 | 104 | 132 | |
| Menopausal status | ||||
| Pre- | 411 | 188 | 223 | 0.041* |
| Post- | 1,493 | 768 | 725 | |
*, P<0.05; **, P<0.01; ***, P<0.001. SET, SE translation.
Table 2
| Characteristics | P value | OR (95% CI) |
|---|---|---|
| Age (≥60 vs. <60) | 0.611 | 1.067 (0.831–1.370) |
| Tumor size (≥5 vs. <5 cm) | 0.090 | 1.550 (0.934–2.570) |
| Histological grade (III vs. I/II) | <0.001*** | 1.535 (1.208–1.950) |
| Tumor stage (III/IV vs. 0/I/II) | 0.240 | 1.301 (0.839–2.019) |
| ER status (positive vs. negative) | <0.001*** | 0.484 (0.348–0.678) |
| PR status (positive vs. negative) | 0.726 | 1.048 (0.807–1.362) |
| Her-2 status (positive vs. negative) | 0.281 | 0.824 (0.579–1.172) |
| Menopausal status (post- vs. pre-) | 0.959 | 0.992 (0.740–1.330) |
***, P<0.001. SET, SE translation.
Potential prognostic factors for breast cancer patients
The univariate COX analysis revealed that diagnosed age (P<0.001), tumor size (P<0.001), neoplasm histologic grade (P<0.001), tumor stage (P<0.001), ER status (P=0.022), PR status (P<0.001), HER-2 status (P<0.001), inferred menopausal status (P<0.001), and SET mRNA expression (P=0.001) influenced the OS of breast cancer patients.
Furthermore, the multivariate COX analysis demonstrated that diagnosed age (P<0.001), tumor histological grade (P=0.018), tumor stage (P<0.001), and HER-2 status (P<0.001) were independent prognostic factors for breast cancer patients (Table 3). The poor prognosis was related to higher ages, poorer tumor tissue differentiation, advanced clinical stage, and positive HER-2 receptor status.
Table 3
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| P value | HR (95% CI) | P value | HR (95% CI) | ||
| Age (≥60 vs. <60) | <0.001*** | 2.120 (1.880–2.389) | <0.001*** | 2.138 (1.813–2.521) | |
| Tumor size (≥5 vs. <5 cm) | <0.001*** | 1.832 (1.499–2.240) | 0.101 | 1.292 (0.951–1.755) | |
| Histological grade (III vs. I/II) | <0.001*** | 1.325 (1.174–1.496) | 0.018* | 1.210 (1.033–1.417) | |
| Tumor stage (III/IV vs. 0/I/II) | <0.001*** | 2.390 (1.925–2.969) | <0.001*** | 1.877 (1.438–2.451) | |
| ER status (positive vs. negative) | 0.022* | 0.848 (0.737–0.977) | 0.589 | 0.944 (0.765–1.164) | |
| PR status (positive vs. negative) | <0.001*** | 0.787 (0.700–0.886) | 0.245 | 0.903 (0.761–1.072) | |
| Her-2 status (positive vs. negative) | <0.001*** | 0.688 (0.579–0.819) | <0.001*** | 1.525 (1.224–1.899) | |
| Menopausal status (post- vs. pre-) | <0.001*** | 1.686 (1.432–1.984) | 0.226 | 1.143 (0.921–1.419) | |
| mRNA expression (high vs. low) | 0.001** | 1.230 (1.092–1.386) | 0.063 | 1.151 (0.992–1.335) | |
*, P<0.05; **, P<0.01; ***, P<0.001.
The effect of SET mRNA expression on the prognosis of breast cancer patients
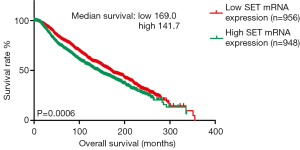
According to Kaplan-Meier analysis, the OS of breast cancer patients with higher SET mRNA expression was significantly poorer than that of patients with lower SET mRNA expression (P=0.006). The result is showed in Figure 2.
Mechanism of SET and its related genes function in breast cancer
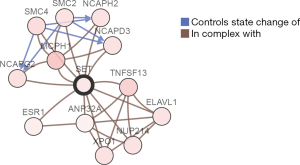
The network searched from cBioPortal website is showed in Figure 3. Total 12 genes related to SET were altered. The regulation of genes is showed in Table 4. Gene interaction types include “control state change of” and “in complex with”.
Table 4
| Gene nodes | Total alteration (%) | Up-regulation (%) | Down-regulation (%) |
|---|---|---|---|
| SET | 3.5 | 2.6 | 1.0 |
| SMC2 | 4.4 | 3.0 | 1.4 |
| SMC4 | 5.2 | 4.3 | 1.0 |
| NCAPG2 | 4.7 | 4.1 | 0.6 |
| NCAPH2 | 4.4 | 2.9 | 1.6 |
| NCAPD3 | 4.5 | 2.5 | 2.0 |
| MCPH1 | 8.3 | 2.7 | 5.7 |
| ESR1 | 2.5 | 0.0 | 2.5 |
| TNFSF13 | 7.0 | 0.7 | 6.3 |
| ANP32A | 2.0 | 1.9 | 0.0 |
| XPO1 | 4.6 | 3.1 | 1.5 |
| NUP214 | 4.4 | 2.5 | 1.9 |
| ELAVL1 | 4.1 | 3.4 | 0.7 |
SET, SE translation.
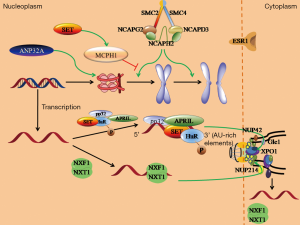
We found that there were two main pathways affecting the progression of breast cancer and patients’ prognosis by analyzing the network and searching literatures. The detail information of signaling pathways can be seen in Figure 4. One pathway was the regulating effect of condensin II on chromosome. As the data displayed, the five subunits – SMC2, SMC4, NCAPG2, NCAPH2, and NCAPD3 of condensin II were upregulated in most breast cancer patients. This complex could bind to chromosome to facilitate its condensation and sister chromatids separation (10). MCHP1 could specifically compete for condensin II binding site on chromosome using one of BRCA1 C-terminal (BRCT) domain which located in N terminus. As a result, condensin II would be suppressed and premature chromosome condensation (PCC) would be saved. When SET bounded to BRCT, the function of MCPH1 would be enhanced. Therefore, SET might be a negative regulator of mitosis entry (11,12) which can block G2/M transition, but it would dysfunction when the SET with mutated C-terminus was overexpressed in COS cells (13). We found MCPH1 was downregulated but SET was upregulated in most breast cancer patients. This phenomenon seemed to be paradoxical. However, there was no recording about mutate data of SET in the database. We hypothesized that if the mutation happened, SET wouldn’t work and the function of MCPH1 was weak. Both of the two genes might not influence condensin II. This process could lead to the occurrence of mitosis entry and breast cancer cell proliferation. On the contrary, if SET was not mutated, the downregulation of MCPH1 and SET regulation effects in other pathways such as overcoming gene repression (5), modifying histone and activating PI3K-Akt pathway (3), would also contribute to breast cancer deterioration. This speculation needs to be further confirmed.
The second pathway was the transportation of tumor cells mRNA towards the extranuclear position. AU-rich elements mRNA (ARE-mRNA) was overexpressed in multiple tumor cells (14). Before ARE-mRNA being transported, phosphorylated ELAVL1 (HuR) would bind to its A-U elements (15) to stabilize the complex together with nuclear phosphoprotein ligand pp32, APRIL, and SET (α, β). Then pp32 and APRIL would combine with the nuclear transport receptor XPO1 (CRM1) to mediate mRNA transportation out of nucleus (16,17). For the mRNA without A-U elements, its extranuclear transportation would be mediated by the evolutionarily conserved, heterodimeric transport factor complex NXF1·NXT1. On the cytoplasmic side of nuclear pore complex (NPC), NXF1·NXT1 would interact with the cytoplasmic filament nucleoporins Gle1, NUP42, and NUP214 to promote its separation with mRNA. These steps could guarantee the direction of mRNA exportation (18). In the process of mRNA nuclear transportation, SET acted to stabilize mRNA and to prevent it from degradation, therefore successful transportation would be guaranteed. According to the data, ELAVL1, SET, XPO1, and NUP214 were all upregulated. This could be beneficial to transport mRNA out of nucleus. Then the translation process of tumor cell and breast cancer development would be accelerated.
We also observed upregulation of ANP32A and downregulation of ESR1. Although ANP32A could suppress solid tumor by preventing histone H3 from acetylation, it was also observed that ANP32A was overexpressed in some tumors. The deficiency of ANP32A would decrease acetylation of histone in tumor cells. Therefore, high expression of ANP32A would promote cell proliferation, survival and colony formation (19). This might be the reason why ANP32A upregulated for breast cancer patients. As a member of nuclear receptor family, ESR1 could bind to enhancers of DNA and then interacted with promoters to regulate the recruitment and activation of RNA polymerase II. Finally, the expression of regulated genes would be upregulated (20). For triple-negative breast cancer cells, other abnormally activated signaling pathway would recover cell proliferation and differentiation abilities to counteract the adverse impact of ESR1 deficiency on tumors. Meanwhile, lack of receptors would lead to deficiency of immunotherapy targets. It would be more difficult to treat breast cancer. All these defects could contribute to patients’ poor prognosis (21).
Discussion
Since SET-CAN fusion gene was found in a case of acute undifferentiated leukemia in 1992 (22), the relationship between SET gene and tumor has gained more and more attention. Many studies have found SET overexpression both in RNA and protein levels in different types of tumors such as leukemia, non-Hodgkin lymphoma, hepatocellular carcinoma, pancreatic carcinoma, metastatic colorectal carcinoma, and breast carcinoma. They also found that the overexpression of SET protein was related to poor prognosis in leukemia (5). To the best of our knowledge, limited research has been done to evaluate the prognostic role of SET in breast cancer. Thus, the aim of the study was to determine biological and prognostic value of SET expression by using bioinformatics databases.
In our study, we found that SET mRNA was significantly associated with diagnosed age, tumor size, neoplasm histologic grade, tumor stage, ER status, PR status, HER-2 status, and inferred menopausal status. Among those characteristics, tumor histological grade and ER status were independent influence factors. Moreover, SET mRNA overexpression could act as a poor prognostic factor for breast cancer patients. These results are in agreement with previous studies (23-25).
There were two pathways involved in the process of breast cancer. They were the facilitation of condensin II on mitosis and the accelerated transportation of tumor cell mRNA towards the extranuclear position. In the first pathway, overexpressed-SET acted to suppress condensin II thereby inhibited mitosis, whereas mutated SET wouldn’t influence condensin II. In the second pathway, SET acted to stabilize mRNA of breast cancer cells. These processes could be regarded as positive factor for tumor cell proliferation. All the conclusions above can be a supplemental demonstration of previous researches (26-37).
Indeed, there are still some limitations in this study. We only analyzed SET mRNA expression for breast cancer patients due to the deficiency of protein expression and mutation data in METABRIC, and the sample size analyzed in this article was not large enough to generate the comprehensive results. These shortcomings can be overcome by conducting researches with more samples in other databases. Besides, further large-size animal experiments and clinical researches are needed to elucidate the role of SET in breast cancer. Finally, as the mechanism of SET was not clearly demonstrated (38-40), other regulating ways of SET can be deeply studied in the future.
Conclusions
SET mRNA tends to overexpress for breast cancer patients with poor tumor tissue differentiation and negative ER status. These patients might have a poorer prognosis. The same clinical outcome would also occur in those with higher ages, poorer tumor tissue differentiation, advanced clinical stage, or positive HER-2 receptor. For the progression of breast cancer, SET can regulate chromosome condensation and stabilize tumor cell mRNA thereby facilitating tumor proliferation and deterioration. These conclusions can be helpful in understanding SET function in breast cancer.
Acknowledgments
We would like to thank the efforts of the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) project.
Funding: The work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.11). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and individual informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Board PDQATE. Breast Cancer Treatment (PDQ(R)): Health Professional Version. National Cancer Institute, Bethesda, 2019.
- Zhang W, Cai J, Chen S, et al. Paclitaxel resistance in MCF-7/PTX cells is reversed by paeonol through suppression of the SET/phosphatidylinositol 3-kinase/Akt pathway. Mol Med Rep 2015;12:1506-14. [Crossref] [PubMed]
- Nagata K, Saito S, Okuwaki M, et al. Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res 1998;240:274-81. [Crossref] [PubMed]
- Bayarkhangai B, Noureldin S, Yu L, et al. A comprehensive and perspective view of oncoprotein SET in cancer. Cancer Med 2018;7:3084-94. [Crossref] [PubMed]
- Arif M, Wei J, Zhang Q, et al. Cytoplasmic retention of protein phosphatase 2A inhibitor 2 (I2PP2A) induces Alzheimer-like abnormal hyperphosphorylation of Tau. J Biol Chem 2014;289:27677-91. [Crossref] [PubMed]
- Jiang SW, Xu S, Chen H, et al. Oncogenic Role of SET/I2PP2A for Gynecologic Cancers. Curr Drug Targets 2017;18:1152-7. [Crossref] [PubMed]
- Fujiki H, Sueoka E, Watanabe T, et al. The concept of the okadaic acid class of tumor promoters is revived in endogenous protein inhibitors of protein phosphatase 2A, SET and CIP2A, in human cancers. J Cancer Res Clin Oncol 2018;144:2339-49. [Crossref] [PubMed]
- Cristóbal I, Rincon R, Manso R, et al. Deregulation of the PP2A inhibitor SET shows promising therapeutic implications and determines poor clinical outcome in patients with metastatic colorectal cancer. Clin Cancer Res 2015;21:347-56. [Crossref] [PubMed]
- Hirano T. Condensins: organizing and segregating the genome. Curr Biol 2005;15:R265-75. [Crossref] [PubMed]
- Yamashita D, Shintomi K, Ono T, et al. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol 2011;194:841-54. [Crossref] [PubMed]
- Leung JW, Leitch A, Wood JL, et al. SET nuclear oncogene associates with microcephalin/MCPH1 and regulates chromosome condensation. J Biol Chem 2011;286:21393-400. [Crossref] [PubMed]
- Canela N, Rodriguez-Vilarrupla A, Estanyol JM, et al. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J Biol Chem 2003;278:1158-64. [Crossref] [PubMed]
- Hitti E, Bakheet T, Al-Souhibani N, et al. Systematic Analysis of AU-Rich Element Expression in Cancer Reveals Common Functional Clusters Regulated by Key RNA-Binding Proteins. Cancer Res 2016;76:4068-80. [Crossref] [PubMed]
- Zhou A, Shi G, Kang GJ, et al. RNA Binding Protein, HuR, Regulates SCN5A Expression Through Stabilizing MEF2C transcription factor mRNA. J Am Heart Assoc 2018;7:1-12. [Crossref] [PubMed]
- Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 2001;294:1895-901. [Crossref] [PubMed]
- Brennan CM, Gallouzi I-E, Steitz JA. Protein Ligands to Hur Modulate Its Interaction with Target Mrnas in Vivo. J Cell Biol 2000;151:1-14. [Crossref] [PubMed]
- Lin DH, Correia AR, Cai SW, et al. Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat Commun 2018;9:2319. [Crossref] [PubMed]
- Yang X, Lu B, Sun X, et al. ANP32A regulates histone H3 acetylation and promotes leukemogenesis. Leukemia 2018;32:1587-97. [Crossref] [PubMed]
- Hah N, Kraus WL. Hormone-regulated transcriptomes: lessons learned from estrogen signaling pathways in breast cancer cells. Mol Cell Endocrinol 2014;382:652-64. [Crossref] [PubMed]
- Neophytou C, Boutsikos P, Papageorgis P. Molecular Mechanisms and Emerging Therapeutic Targets of Triple-Negative Breast Cancer Metastasis. Front Oncol 2018;8:31. [Crossref] [PubMed]
- von Lindern M, van Baal S, Wiegant J, et al. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3' half to different genes: characterization of the set gene. Mol Cell Biol 1992;12:3346-55. [Crossref] [PubMed]
- Yuan X, Zhang T, Zheng X, et al. Overexpression of SET oncoprotein is associated with tumor progression and poor prognosis in human gastric cancer. Oncol Rep 2017;38:1733-41. [Crossref] [PubMed]
- Lin Z, Jia H, Hong L, et al. Prognostic impact of SET domain-containing protein 8 and protein arginine methyltransferase 5 in patients with hepatocellular carcinoma following curative resection. Oncol Lett 2018;16:3665-73. [PubMed]
- Liu H, Gu Y, Wang H, et al. Overexpression of PP2A inhibitor SET oncoprotein is associated with tumor progression and poor prognosis in human non-small cell lung cancer. Oncotarget 2015;6:14913-25. [PubMed]
- Wang HZ, Yang SH, Li GY, et al. Subunits of human condensins are potential therapeutic targets for cancers. Cell Div 2018;13:2. [Crossref] [PubMed]
- Kagami Y, Yoshida K. The functional role for condensin in the regulation of chromosomal organization during the cell cycle. Cell Mol Life Sci 2016;73:4591-8. [Crossref] [PubMed]
- Zhang C, Kuang M, Li M, et al. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci Rep 2016;6:34508. [Crossref] [PubMed]
- Shiheido H, Naito Y, Kimura H, et al. An anilinoquinazoline derivative inhibits tumor growth through interaction with hCAP-G2, a subunit of condensin II. PloS one 2012;7:e44889. [Crossref] [PubMed]
- Lapointe J, Malhotra S, Higgins JP, et al. hCAP-D3 expression marks a prostate cancer subtype with favorable clinical behavior and androgen signaling signature. Am J Surg Pathol 2008;32:205-9. [Crossref] [PubMed]
- Wang Y, He PC, Liu YF, et al. Construction of SET overexpression vector and its effects on the proliferation and apoptosis of 293T cells. Mol Med Rep 2016;13:4329-34. [Crossref] [PubMed]
- Hung MH, Chen KF. Reprogramming the oncogenic response: SET protein as a potential therapeutic target in cancer. Expert Opin Ther Targets 2017;21:685-94. [Crossref] [PubMed]
- Wu M, Tong CWS, Yan W, et al. The RNA Binding Protein HuR: a Promising Drug Target for Anticancer Therapy. Curr Cancer Drug Targets 2019;19:382-99. [Crossref] [PubMed]
- Heinonen M, Fagerholm R, Aaltonen K, et al. Prognostic role of HuR in hereditary breast cancer. Clin Cancer Res 2007;13:6959-63. [Crossref] [PubMed]
- Gravina GL, Senapedis W, McCauley D, et al. Nucleo-cytoplasmic transport as a therapeutic target of cancer. J Hematol Oncol 2014;7:85. [Crossref] [PubMed]
- Conforti F, Wang Y, Rodriguez JA, et al. Molecular Pathways: Anticancer Activity by Inhibition of Nucleocytoplasmic Shuttling. Clin Cancer Res 2015;21:4508-13. [Crossref] [PubMed]
- Cruz-Ramos E, Sandoval-Hernandez A, Tecalco-Cruz AC. Differential expression and molecular interactions of chromosome region maintenance 1 and calreticulin exportins in breast cancer cells. J Steroid Biochem Mol Biol 2019;185:7-16. [Crossref] [PubMed]
- Luan X, Wang Y. Long non-coding RNA XLOC_006390 promotes cervical cancer proliferation and metastasis through the regulation of SET domain containing 8. Oncol Rep 2017;38:159-66. [Crossref] [PubMed]
- Yuan X, Wang X, Gu B, et al. Directional Migration in Esophageal Squamous Cell Carcinoma (ESCC) is Epigenetically Regulated by SET Nuclear Oncogene, a Member of the Inhibitor of Histone Acetyltransferase Complex. Neoplasia 2017;19:868-84. [Crossref] [PubMed]
- Almeida LO, Garcia CB, Matos-Silva FA, et al. Accumulated SET protein up-regulates and interacts with hnRNPK, increasing its binding to nucleic acids, the Bcl-xS repression, and cellular proliferation. Biochem Biophys Res Commun 2014;445:196-202. [Crossref] [PubMed]