
The high expression of lncRNA TUG1 correlates with progressive tumor condition and less satisfying survival profiles in prostate cancer patients
Introduction
Prostate cancer is the most frequent malignancy troubling men worldwide, exhibiting an escalating incidence and causing an estimated 603,000 new cancer cases and 266,000 cancer-related deaths in China in 2015 (1,2). The diagnosis of prostate cancer has faced a dilemma over the past few decades, stemming from an invasive diagnostic procedure that usually performs biopsy through the entire organ, along with overdiagnosis caused by screening based on a prostate-specific antigen (PSA) (3,4). Also, standard treatment modalities of prostate cancer are also problematic, and predominantly include prostatectomy, radiotherapy, brachytherapy for localized disease, and anti-testicular androgen treatments for metastatic disease. These are further hampered by a lack of investigation by randomized controlled trials and the expensive nature fee of promising novel drugs (5-11). Therefore, the discovery of potential prognostic biomarkers is urgently needed.
Long noncoding RNAs (lncRNAs), a new category of RNA with no coding capability and with a length of more than 200 nt, have been found by emerging studies to have the ability to mediate gene expression through combining with proteins, other RNA, or DNA at both the transcriptional and post-transcriptional level (12-14). lncRNA taurine-upregulated 1 gene (TUG1) is a promising lncRNA being focused on in oncology research and has shown to be correlated with the progression of various solid tumors, such as melanoma and bladder cancer (15,16). However, despite abundant studies reporting its oncogenic role in tumorigenesis, the investigation of lncRNA TUG1 in prostate cancer is extremely insufficient. A previous study found the potential anti-tumorigenesis function of lncRNA TUG1 in prostate cancer through microarray analysis, but the results largely conflict with previous findings (17). Moreover, the sample size in this study was small, and there could have been some inconsistency in the microarray analysis. Thus, our study aimed to investigate the correlation between lncRNA TUG1 and tumor characteristics and survival in prostate cancer patients, along with the effect of lncRNA TUG1 on cell proliferation and cell apoptosis in prostate cancer cells.
Methods
Patients
In total, 208 prostate cancer patients who underwent tumor resection were continuously reviewed in this study between January 1st, 2012, and December 31st, 2015, in The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. Patients with the following conditions were included: (I) pathologically diagnosed as primary prostate cancer; (II) age above 18 years; (III) complete cancer features at surgery which at least included PSA level, Gleason score, pathological stage, and lymph node metastasis status; (IV) regularly followed up with complete disease-free survival (DFS) and overall survival (OS) data; (V) fresh tumor tissue and paired adjacent tissue stored in liquid nitrogen in the Specimen Store House. Meanwhile, patients with previous prostate surgery, previous other solid tumors or malignant hematological diseases, recurrent prostate cancer or received neo-adjuvant therapy, were excluded. The study was approved by the Human Ethics Review Board of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology of No. 2016-3, and written informed consent was obtained from all participants.
Sample collection and quantitative polymerase chain reaction (qPCR) assay
Tumor tissue and paired adjacent tissue collected during surgery were obtained from the Specimen Store House, and qPCR assays were performed to detect the expression of lncRNA TUG1 (LncRNA TUG1 (ENSG00000253352) forward: CTCTTGCCTGCCTTTTTCAC; reverse: GCAATGGTCCTTCCTGTTGT). Total RNA was extracted by using TRIzol solution (Invitrogen, USA) strictly following the instructions of the manufacturer. Total RNA was then reverse transcribed by PrimerScript Real-time reagent kit (TaKaRa, Japan), and labeled by KAPA SYBR FAST qPCR kit (KAPA, USA). Subsequently, glyceraldehyde-phosphate dehydrogenase (GAPDH) was used as an internal reference of the lncRNA TUG1 expression standardization, which was finally calculated using the 2−∆∆t method (GAPDH forward: 5'-TGACCACAGTCCATGCCATCAC-3'; reverse: 5'-GCCTGCTTCACCACCTTCTTGA-3').
Data collection, follow-up, and survival profiles
Patient features were collected including age, PSA level, Gleason score, pathological stage, lymph node metastasis status, and surgical margin status. Patients were regularly followed up with, and the median duration was 40 months (the last follow-up occurred on December 31st, 2017), and DFS and OS were calculated.
Cell culture
The PANC-1 cells, which were purchased from Cell Resource Center of Shanghai Institute of Life Sciences (Shanghai, China), were cultured in a 90% DMEM medium (Gibco, USA) which was supplemented with 10% fetal bovine serum (FBS) (Gibco, USA). The cells were then put in a humidified incubator in a 95% air and 5% CO2 environment at 37 °C.
Transfections and subsequent assays
Control overexpression, lncRNA TUG1 overexpression, control shRNA, and lncRNA TUG1 shRNA plasmids were constructed and transfected into the PANC-1 cells as the NC(+) group, TUG1(+) group, NC(−) group, and TUG1(−) group, respectively. Cell counting kit-8 (CCK-8) assays were performed at 0, 24, 48, and 72 h to assess the cell proliferation, and an AV/PI assay was performed at 24 h to detect cell apoptosis in PANC-1 cells.
CCK-8
CCK-8 was used to evaluate the cell proliferation ability. First, 10 µL CCK-8 (Abcam, USA) and 90 µL medium were added to each plate, and the cells were incubated under 95% air and 5% CO2 at 37 °C. After this, the optical density (OD) value was assessed by a microplate reader (BioTeck, USA) for the measurement of proliferation ability.
AV/PI
The cells were digested and washed, after which 2 µL AV (Invitrogen, USA) and 1 µL PI (Invitrogen, USA) were added to each plate, which was placed in darkness for 15 min. Finally, the apoptosis rate was detected by the flow cytometry (Becton Dickinson, USA) following the instructions of the manufacturer.
Statistics
SPSS 22.0 software (IBM, USA) was used for statistical analysis, and GraphPad Prism (GraphPad Inc., USA) was applied for graph making. Data were mainly presented as mean ± standard deviation, median (1/4–3/4 quantiles), and count (percentage). The comparison was determined by Wilcoxon signed rank-sum test, Wilcoxon rank-sum test, or t-test; comparison of DFS and OS between the two groups was determined by Kaplan-Meier (K-M) curve and log-rank test; factors for predicting DFS and OS were analyzed with univariate and multivariate Cox’s proportional hazards model. P<0.05 was considered statistically significant.
Results
Baseline characteristics of prostate cancer patients
As listed in Table 1, the mean age of our 208 prostate cancer patients was 61.7±9.0 years. The numbers of patients with PSA level ≤10, 10–20, and ≥20 ng/mL were 60 (28.8%), 103 (49.6%), and 45 (21.6%), respectively. In addition, there were 46 (22.1%), 114 (54.8%), and 48 (23.1%) patients with Gleason score ≤6, 7, and ≥8, respectively. The numbers of patients with pathological stage pT2a, pT2b, pT2c, pT3a, pT3b and pT4 were 59 (28.4%), 37 (17.8%), 28 (13.5%), 39 (18.8%), 41 (19.7%), and 4 (1.9%), respectively. Sixty (28.8%) patients had lymph node metastasis while 148 (71.2%) patients had no lymph node metastasis. The numbers of patients with positive and negative surgical margin status were 59 (28.4%) and 149 (71.6%), respectively.
Table 1
| Parameters | Prostate cancer patients (N=208) |
|---|---|
| Age (years) | 61.7±9.0 |
| PSA (n/%) | |
| ≤10 ng/mL | 60 (28.8) |
| 10–20 ng/mL | 103 (49.6) |
| ≥20 ng/mL | 45 (21.6) |
| Gleason score (n/%) | |
| ≤6 | 46 (22.1) |
| 7 | 114 (54.8) |
| ≥8 | 48 (23.1) |
| Pathological stage (n/%) | |
| pT2a | 59 (28.4) |
| pT2b | 37 (17.8) |
| pT2c | 28 (13.5) |
| pT3a | 39 (18.8) |
| pT3b | 41 (19.7) |
| pT4 | 4 (1.9) |
| Lymph node metastasis (n/%) | |
| No | 148 (71.2) |
| Yes | 60 (28.8) |
| Surgical margin status (n/%) | |
| Negative | 149 (71.6) |
| Positive | 59 (28.4) |
Data were presented as mean ± standard deviation or count (%). PSA, prostate-specific antigen.
Expression of lncRNA TUG1 in tumor tissue and paired adjacent tissue
The expressions of lncRNA TUG1 in tumor tissue and paired adjacent tissue were detected by qPCR, which showed that lncRNA TUG1 expression was elevated in tumor tissue compared with paired adjacent tissue (P<0.001) (Figure 1).
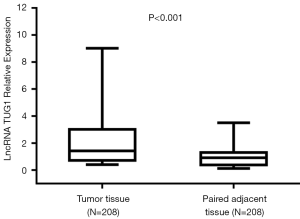
Correlation of lncRNA TUG1 expression in tumor tissue with tumor characteristics
As shown in Table 2, lncRNA TUG1 expression in tumor tissue positively correlated with pathological stage (P=0.009) and lymph node metastasis (P=0.040), while it was not associated with age (P=0.716), PSA level (P=0.443), Gleason score (P=0.345) or surgical margin status (P=0.863) in prostate cancer patients.
Table 2
| Parameters | LncRNA TUG1 relative expression | P value |
|---|---|---|
| Age | 0.716 | |
| <60 years | 1.465 (0.660–3.009) | |
| ≥60 years | 1.416 (0.763–3.039) | |
| PSA | 0.443 | |
| ≤10 ng/mL | 1.637 (0.675–3.396) | |
| 10–20 ng/mL | 1.357 (0.762–2.913) | |
| ≥20 ng/mL | 1.306 (0.630–2.247) | |
| Gleason score | 0.345 | |
| ≤6 | 1.544 (0.779–2.836) | |
| 7 | 1.329 (0.614–2.842) | |
| ≥8 | 1.638 (0.798-3.840) | |
| Pathological stage | 0.009 | |
| pT2 | 1.130 (0.660–2.634) | |
| pT3 | 1.917 (0.944–3.794) | |
| pT4 | 3.784 (1.221–5.124) | |
| Lymph node metastasis | 0.040 | |
| No | 1.258 (0.702–2.723) | |
| Yes | 1.942 (0.730–4.623) | |
| Surgical margin status | 0.863 | |
| Negative | 1.446 (0.696–2.941) | |
| Positive | 1.349 (0.763–3.258) | P value |
Data were presented as median (25th–75th quantiles). Comparison was determined by Wilcoxon rank sum test or Kruskal-Wallis H rank sum test. P value <0.05 was considered as significant. PSA, prostate-specific antigen.
Correlation of lncRNA TUG1 expression in tumor tissue with DFS and OS
As displayed in Figure 2, prostate cancer patients with high lncRNA TUG1 expression had both worse DFS (P<0.001) (Figure 2A) and OS (P=0.003) (Figure 2B) compared with patients with low lncRNA TUG1 expression.
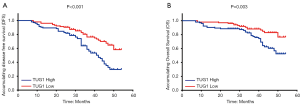
Predictive value of lncRNA TUG1 expression for DFS and OS
To evaluate the predictive value of lncRNA TUG1 expression in tumor tissue for DFS and OS, we performed univariate and multivariate Cox’s proportional hazards regression analyses. The univariate Cox’s proportional hazards regression analysis disclosed that high lncRNA TUG1 expression (P<0.001), higher Gleason score (P=0.001), higher pathological stage (P<0.001), and positive surgical margin status (P=0.014) correlated with shorter DFS (Table 3). Subsequently, all factors were included in the multivariate Cox’s proportional hazards regression analysis, which revealed that high lncRNA TUG1 expression (P=0.001), higher PSA (P=0.026), higher Gleason score (P<0.001), higher pathological stage (P<0.001), and positive surgical margin status (P=0.008) were independent predictive factors for worse DFS.
Table 3
| Parameters | Univariate Cox’s regression | Multivariate Cox’s regression | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | ||||
| Lower | Higher | Lower | Higher | ||||||
| LncRNA TUG1 high expression | <0.001 | 2.285 | 1.455 | 3.588 | 0.001 | 2.265 | 1.423 | 3.605 | |
| Age (≥60 years) | 0.846 | 0.958 | 0.622 | 1.477 | 0.941 | 1.017 | 0.655 | 1.579 | |
| Higher PSA | 0.325 | 1.166 | 0.859 | 1.585 | 0.026 | 1.431 | 1.045 | 1.960 | |
| Higher Gleason score | 0.001 | 1.828 | 1.294 | 2.580 | <0.001 | 1.967 | 1.387 | 2.789 | |
| Higher pathological stage | <0.001 | 2.087 | 1.408 | 3.093 | <0.001 | 2.107 | 1.416 | 3.136 | |
| Lymph node metastasis (yes) | 0.101 | 1.491 | 0.925 | 2.404 | 0.224 | 1.364 | 0.827 | 2.250 | |
| Surgical margin status (positive) | 0.014 | 1.775 | 1.122 | 2.809 | 0.008 | 1.889 | 1.184 | 3.014 | |
Data were presented as P value, HR and 95% CI. PSA stage was scored as: ≤10 ng/mL =0, 10–20 ng/mL =1, ≥20 ng/mL =2. Gleason score was scored as: ≤6 point =0, 7 point =1, ≥8 point =2. Pathological stage was scored as: pT2=2, pT3=3, pT4=4. P value <0.05 was considered significant. DFS, disease free survival; PSA, prostate-specific antigen; HR, hazards ratio; CI, confidence interval.
As for OS, the results of univariate Cox’s proportional hazards regression analysis showed that high lncRNA TUG1 expression (P=0.004), higher Gleason score (P=0.008), and positive surgical margin status (P=0.039) were predicting factors for less favorable OS (Table 4). Subsequently, all the factors were analyzed by multivariate Cox’s proportional hazards regression analysis, which exhibited that high lncRNA TUG1 expression (P=0.003), higher PSA (P=0.006), higher Gleason score (P=0.002), higher pathological grade (P=0.043), and positive surgical margin status (P=0.019) were independently associated with poorer OS.
Table 4
| Parameters | Univariate Cox’s regression | Multivariate Cox’s regression | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | ||||
| Lower | Higher | Lower | Higher | ||||||
| LncRNA TUG1 high expression | 0.004 | 2.448 | 1.329 | 4.509 | 0.003 | 2.555 | 1.362 | 4.794 | |
| Age (≥60 years) | 0.319 | 1.351 | 0.747 | 2.443 | 0.277 | 1.396 | 0.765 | 2.546 | |
| Higher PSA | 0.110 | 1.395 | 0.928 | 2.099 | 0.006 | 1.810 | 1.190 | 2.753 | |
| Higher Gleason score | 0.008 | 1.867 | 1.175 | 2.966 | 0.002 | 2.104 | 1.313 | 3.371 | |
| Higher pathological stage | 0.061 | 1.661 | 0.976 | 2.827 | 0.043 | 1.743 | 1.019 | 2.980 | |
| Lymph node metastasis (yes) | 0.057 | 1.828 | 0.981 | 3.407 | 0.087 | 1.778 | 0.921 | 3.433 | |
| Surgical margin status (positive) | 0.039 | 1.873 | 1.031 | 3.403 | 0.019 | 2.080 | 1.130 | 3.828 | |
Data were presented as P value, HR and 95% CI. PSA stage was scored as: ≤10 ng/mL =0, 10–20 ng/mL =1, ≥20 ng/mL =2. Gleason score was scored as: ≤6 point =0, 7 point =1, ≥8 point =2. Pathological stage was scored as: pT2=2, pT3=3, pT4=4. P value <0.05 was considered significant. OS, overall survival; PSA, prostate-specific antigen; HR, hazards ratio; CI, confidence interval.
Effect of lncRNA TUG1 on cell proliferation and apoptosis in prostate cancer cells
After the transfections, the cell proliferation at 72 h was enhanced in the TUG1(+) group compared with the NC(+) group (P<0.05) while it was decreased in the TUG1(−) group compared with the NC(−) group (P<0.05) (Figure 3A). As for cell apoptosis, the cell apoptosis rate declined in the TUG1(+) group compared with the NC(+) group (P<0.05) while it was elevated in the TUG1(−) group compared with the NC(−) group (P<0.01) (Figure 3B,C).
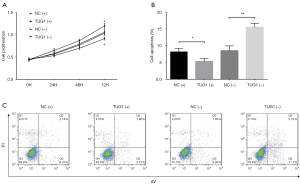
Discussion
In our study, we discovered that (I) lncRNA TUG1 was upregulated in tumor tissue, which was also correlated with higher pathological stage and lymph node metastasis in prostate cancer patients; (II) prostate cancer patients with high lncRNA TUG1 expression in tumor tissue had both shorter DFS and OS compared to patients with low lncRNA TUG1 expression. Multivariate Cox’s proportional hazards regression analysis determined that high lncRNA expression was an independent predicting factor for both worse DFS and OS; (III) lncRNA TUG1 promoted cell proliferation while suppressing cell apoptosis in prostate cancer cells.
LncRNA TUG1, which is abundantly expressed in normal tissue and tumor cells, is originally discovered in murine retinal cells, can be upregulated by taurine, and is also found to be crucial to photoreceptor formation (18). In recent years, the number of studies reporting lncRNA TUG1 to be a tumor-related lncRNA has continued to increase, while the study evidence for its pro- or anti-tumorigenesis has also progressively accumulated. A prior in vitro experiment elucidates that repressing the expression of lncRNA TUG1 inhibits cells growth and metastasis via mediating the astrocyte-elevated gene-1 expression by targeting miR-129-5p in melanoma (15). Another experiment revealed that lncRNA TUG1 enhances cells proliferation while repressing cell apoptosis via sponging miR-132-3p in osteosarcoma (19). Also, knocking down lncRNA TUG1 also suppresses the proliferation, motility, and invasiveness of intrahepatic cholangiocarcinoma cells through Sirt3/GDH axis (20).
Moreover, upregulating lncRNA TUG1 in bladder cancer cells boosts cell proliferation, migration, and invasion via suppressing miR-29c (16). In our study, we found that lncRNA TUG1 was upregulated in tumor tissue and positively correlated with pathological stage and lymph node metastasis in prostate cancer patients. These results from investigations with large-sample-size populations provide new insight into the role of lncRNA TUG1 in prostate cancer. The possible explanation for our results might be that lncRNA TUG1, as indicated by these previous studies, may have the ability to promote tumor progression by enhancing tumor cell proliferation, growth, and metastasis while inhibiting tumor cell apoptosis through sponging several downstaging miRNAs including miR-132-3p and miR-29c, and regulating pathways such as Sirt3/GDH axis and TUG1/Mir-129-5p/Astrocyte-Elevated Gene-1 (AEG-1) axis (15,16,19,20). More importantly, we further conducted cell experiments in our study and found that lncRNA TUG1 enhanced cell proliferation while inhibiting cell apoptosis in prostate cancer cells, which also provided valuable information on the molecular mechanism of the carcinogenic role that lncRNA TUG1 plays in prostate cancer.
Given its great potential for cancer treatment, the prognostic role of lncRNA TUG1 in cancers is also fully investigated. One meta-analysis suggests that elevated lncRNA TUG1 expression in tumor tissue notably correlates with poorer OS in patients with multiple cancers (21). For instance, the high expression of lncRNA TUG1 in tumor tissue associates with worse OS in high-grade muscle-invasive bladder cancer patients (22). Meanwhile, increased lncRNA TUG1 expression in tumor tissue is an independent predictive factor for poorer OS in clear cell renal cell carcinoma patients (23). It has also been reported that small cell lung cancer patients with higher tumor tissue expression of lncRNA TUG1 have shorter OS compared to those with a lower expression of lncRNA TUG1 in tumor tissue (24). Also, they also elucidate that lncRNA TUG1 has a function of increasing the chemoresistance of small cell lung cancer cells by modulating LIMK2b through binding with the enhancer of zeste homolog 2 (24). Similar to the previous studies, prostate cancer patients in our study who had higher expressions of lncRNA TUG1 in tumor tissue suffered worse DFS and OS. More importantly, our multivariate Cox’s proportional hazards regression analysis revealed that high lncRNA TUG1 expression in tumor tissue was an independent predictive factor for both worse DFS and OS. It is probably due to lncRNA enhancing cell proliferation and migration while repressing the cell apoptosis of tumor cells via sponging multiple downstaging miRNAs which include miR-132-3p and miR-29c and mediating pathways like Sirt3/GDH axis. lncRNA may also increase drug resistance in treatment areas like chemotherapeutics, which could subsequently lead to the promotion of tumor progression and result in a worse survival (15,16,19,20,24).
There were several limitations to our study. First, we evaluated the expression of lncRNA TUG1 only in tumor tissue but not in circulating samples of prostate cancer patients; lnc TUG1tumor expression cannot be applied as a biomarker for disease monitoring. However, there is still no evidence suggesting that lncRNA TUG1 can be detected in circulating samples of prostate cancer patients; therefore, we only evaluated its expression level in tumor tissue. Second, the specific mechanism of lncRNA TUG1 promoting progression of prostate cancer was not investigated in our study in detail, and this phenomenon should be validated by further in vivo and in vitro experiments.
In conclusion, lncRNA TUG1 high expression in tumor tissue correlates with progressive tumor condition and poorer survival in prostate cancer patients and promotes proliferation while inhibits cell apoptosis in prostate cancer cells.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Human Ethics Review Board of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology of No. 2016-3. And all patients or their guardians signed the informed consents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Cancer risk. Cancer Research UK. 2011. Available online: http://www.cancerresearchuk.org/cancer-info/cancerstats/incidence/risk/
- Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet 2016;387:70-82. [Crossref] [PubMed]
- Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015;313:390-7. [Crossref] [PubMed]
- Bill-Axelson A, Holmberg L, Ruutu M, et al. Radical prostatectomy versus watchful waiting in early prostate cancer. N Engl J Med 2011;364:1708-17. [Crossref] [PubMed]
- Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med 2012;367:203-13. [Crossref] [PubMed]
- Widmark A, Klepp O, Solberg A, et al. Endocrine treatment, with or without radiotherapy, in locally advanced prostate cancer (SPCG-7/SFUO-3): an open randomised phase III trial. Lancet 2009;373:301-8. [Crossref] [PubMed]
- Warde P, Mason M, Ding K, et al. Combined androgen deprivation therapy and radiation therapy for locally advanced prostate cancer: a randomised, phase 3 trial. Lancet 2011;378:2104-11. [Crossref] [PubMed]
- Horwitz EM, Bae K, Hanks GE, et al. Ten-year follow-up of radiation therapy oncology group protocol 92-02: a phase III trial of the duration of elective androgen deprivation in locally advanced prostate cancer. J Clin Oncol 2008;26:2497-504. [Crossref] [PubMed]
- Denham JW, Steigler A, Lamb DS, et al. Short-term neoadjuvant androgen deprivation and radiotherapy for locally advanced prostate cancer: 10-year data from the TROG 96.01 randomised trial. Lancet Oncol 2011;12:451-9. [Crossref] [PubMed]
- Maximum androgen blockade in advanced prostate cancer: an overview of the randomised trials. Prostate Cancer Trialists' Collaborative Group. Lancet 2000;355:1491-8. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res 2015;25:1456-65. [Crossref] [PubMed]
- Long J, Menggen Q, Wuren Q, et al. Long Noncoding RNA Taurine-Upregulated Gene1 (TUG1) Promotes Tumor Growth and Metastasis Through TUG1/Mir-129-5p/Astrocyte-Elevated Gene-1 (AEG-1) Axis in Malignant Melanoma. Med Sci Monit 2018;24:1547-59. [Crossref] [PubMed]
- Guo P, Zhang G, Meng J, et al. Upregulation of Long Noncoding RNA TUG1 Promotes Bladder Cancer Cell Proliferation, Migration, and Invasion by Inhibiting miR-29c. Oncol Res 2018;26:1083-91. [Crossref] [PubMed]
- Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun 2016;7:10982. [Crossref] [PubMed]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005;15:501-12. [Crossref] [PubMed]
- Li G, Liu K, Du X. Long Non-Coding RNA TUG1 Promotes Proliferation and Inhibits Apoptosis of Osteosarcoma Cells by Sponging miR-132-3p and Upregulating SOX4 Expression. Yonsei Med J 2018;59:226-35. [Crossref] [PubMed]
- Zeng B, Ye H, Chen J, et al. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget 2017;8:113650-61. [Crossref] [PubMed]
- Zhou Y, Lu Y, Li R, et al. Prognostic role of long non-coding RNA TUG1 expression in various cancers: a meta-analysis. Oncotarget 2017;8:100499-507. [PubMed]
- Iliev R, Kleinova R, Juracek J, et al. Overexpression of long non-coding RNA TUG1 predicts poor prognosis and promotes cancer cell proliferation and migration in high-grade muscle-invasive bladder cancer. Tumour Biol 2016;37:13385-90. [Crossref] [PubMed]
- Wang PQ, Wu YX, Zhong XD, et al. Prognostic significance of overexpressed long non-coding RNA TUG1 in patients with clear cell renal cell carcinoma. Eur Rev Med Pharmacol Sci 2017;21:82-6. [PubMed]
- Niu Y, Ma F, Huang W, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 2017;16:5. [Crossref] [PubMed]


