
Proteomics score: a potential biomarker for the prediction of prognosis in non-small cell lung cancer
Introduction
Lung cancer is the most common cancer and the leading cause of cancer-related death worldwide. The most prevalent cause of lung cancer mortality, accounting for about 85% of related deaths, is non-small cell lung cancer (NSCLC) (1-3). Although lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), as two major histological subtypes, have distinct biological behaviors and require different therapeutic approaches, pneumonectomy is a potential curative therapy in some patients with LUAD or LUSC (4-6). Approximately 60% of patients with early- to mid-stage NSCLC will never experience recurrence after surgical treatment, whereas 40% eventually die of the disease (7). Therefore, screening for patients with resectable NSCLC at higher risk of death or relapse may help to improve treatment outcomes.
Recently, large-scale genomic analysis techniques, including microarrays and RNA-Seq, have been used to obtain genome-wide mRNA expression data in different types of cancers (8-13). The mRNA expression signatures have been used to predict the prognosis of patients with NSCLC in several studies (14-18). However, the robustness, reproducibility, and clinical applications of these prognostic gene signatures are still unclear. Therefore, determination of which prognostic biomarkers are suitable for extensive and long-term prospective clinical trials is urgently required. Moreover, reliable and novel prognostic biomarkers need to be explored to establish improved clinical therapies.
Functional proteomics is mainly focused on the study of protein activity levels (e.g., expression and modifications) (19,20). In addition to genetic alterations, abnormalities in protein expression levels and structures also play key roles in tumor development and progression. Similar to western blotting, reverse phase protein arrays (RPPAs) are a high-throughput, antibody-based, robust quantification technique that can be used to accurately determine protein levels in various types of tumors (21,22). Indeed, proteomics results are significantly associated with prognosis in many cancers (23,24). However, novel models based on multiple proteomics to predict and validate overall and disease-free survival (OS and DFS, respectively) have not yet been reported in NSCLC cohorts.
Therefore, in this study, we attempted to build a model based on functional proteomics for predicting prognosis in patients with NSCLC after surgery. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to obtain an in-depth understanding of selection proteomics. Gene set enrichment analysis (GSEA) and protein/protein interaction network (PPI) analysis were used to confirm the potential mechanisms.
Methods
Public data acquisition
The National Genome Research Institute and the National Cancer Institute collected tissue samples from The Cancer Genome Atlas (TCGA). Informed consent and ethical approval were obtained. This retrospective study was approved by our institutional review board and Ethical Committee (NFEC-201208-K3). Level 3 data from RPPA and clinical data for LUAD and LUSC were downloaded from cBioPortal (http://www.cbioportal.org). Detailed information regarding protein markers, including the corresponding genes, validation status, and source of antibody, is shown in The Cancer Proteome Atlas (http://tcpaportal.org/tcpa). Tissue microarray (TMA) was selected from the Human Protein Atlas Network (HPAS; https://www.proteinatlas.org). Immunohistochemistry (IHC) scores were evaluated by rescoring of intensity and quantity.
Feature selection and proteomics score development
All patients with LUAD and LUSC were randomly divided into the discovery and validation sets at a 1:1 ratio. Using the LASSO-COX algorithm, we built a model and selected the λ in the smallest cross-validation error in the discovery set. Finally, a proteomics score formula was defined based on the selected features. A proteomics signature was then constructed using the proteomics score. Using Kaplan-Meier survival analysis, we evaluated the potential relationships between the proteomics score and prognosis (OS and DFS) in the discovery and validation sets. The optimal cut-off value for the proteomics score was determined using X-tile software (version 3·6·1) in OS analysis.
Development and validation of the proteomics nomogram
Multivariate Cox regression analysis was performed to build a proteomics nomogram as a quantitative model to predict OS. Candidate predictors of OS were proteomics score and clinical data. The performance of the nomogram was estimated in the two cohorts. OS was then evaluated considering the total points as a factor in the Cox regression analysis. Finally, the C-index and calibration curves were derived using Cox regression analysis. Harrell’s C-index was evaluated to quantify the discrimination capability of the proteomics nomogram in the discovery set. The proteomics nomogram was then validated using 1,000 bootstrap samples to achieve an optimism-corrected performance. Time-dependent receiver operator characteristic (ROC) curves were used to evaluate the predictive accuracy as the area under the ROC curve (AUC) at 1, 3, and 5 years.
Statistical analysis
All statistical analyses were performed using R statistical software version 3.5.0 (http://www.r-project.org) and GraphPad Prism 7.0 (https://www.graphpad.com). The clinical characteristics in the discovery and validation cohorts were analyzed by Chi-square tests. The “glmnet” package was used to perform the LASSO algorithm. The nomogram and calibration curve plot were created using the “rms” package. The results of the Kaplan-Meier survival analysis were plotted using the “survminer” package, whereas those of the time-dependent ROC were plotted using the “survivalROC” package. IHC scores were compared by Mann-Whitney tests. The correlation analysis was performed by Pearson’s correlation analysis. A two-sided P value of less than 0.05 was considered statistically significant.
In bioinformation research, all selection genomics and components of proteomics scores were further analyzed using GO analysis and annotated to pathways using the KEGG database (data generated from DAVID and String; DAVID, https://david.ncifcrf.gov/; String, https://string-db.org/cgi/input.pl). GSEA was used to determine the potential mechanisms of specific genes. The association between PPI and GO/KEGG analyses were visualized using Cytoscape ClueGO version 3.6.1 (http://www.cytoscape.org/).
Results
Clinical characteristics
Six hundred ninety-three patients were included in this study, of whom 346 patients were allocated to the discovery set and 347 were allocated to the validation set. LUAD and LUSC cohorts included 365 and 328 patients, respectively. The baseline clinical characteristics of the discovery and validation sets are summarized in Table 1. Most patients with NSCLC had early- or mid-stage disease (stages I–II) in both cohorts (78.03% and 80.12%, respectively). There were no significant differences between the discovery and validation sets (P=0.087–0.988; Table 1).
Table 1
| Variable | Discovery set, n=346 (%) | Validation set, n=347 (%) | P value |
|---|---|---|---|
| Sex | 0.336 | ||
| Female | 144 (41.62) | 132 (38.04) | |
| Male | 202 (58.38) | 215 (61.96) | |
| Age (years) | 0.408 | ||
| ≤60 | 90 (26.01) | 100 (28.82) | |
| >60 | 256 (73.99) | 247 (71.18) | |
| Smoking indicator | 0.341 | ||
| ≤3 | 187 (54.50) | 200 (57.64) | |
| >3 | 159 (45.50) | 147 (42.36) | |
| ECOG score | 0.695 | ||
| ≤2 | 147 (42.49) | 138 (39.77) | |
| >2 | 4 (1.15) | 3 (0.86) | |
| NA | 195 (56.36) | 206 (59.37) | |
| History of other malignancy | 0.988 | ||
| Yes | 50 (14.45) | 50 (14.41) | |
| No | 296 (85.55) | 297 (85.59) | |
| Histological subtype | 0.087 | ||
| Lung adenocarcinoma | 171 (49.42) | 194 (55.91) | |
| Lung squamous cell carcinoma | 175 (50.58) | 153 (44.09) | |
| Outcome of first treatment | 0.826 | ||
| CR | 84 (24.28) | 78 (22.48) | |
| SD + PD | 15 (4.33) | 14 (4.03) | |
| NA | 247 (71.39) | 255 (73.49) | |
| Width of invasion | 0.822 | ||
| T1 + T2 | 290 (83.82) | 293 (84.44) | |
| T3 + T4 | 56 (16.18) | 54 (15.56) | |
| Lymph node metastasis | 0.459 | ||
| N0 | 216 (62.43) | 226 (65.13) | |
| N1 + N2 + N3 | 130 (37.57) | 121 (34.87) | |
| Distant metastasis | 0.655 | ||
| M0 | 337 (97.40) | 336 (96.83) | |
| M1a + M1b | 9 (2.60) | 11 (3.17) | |
| Clinical stage | 0.501 | ||
| I + II | 270 (78.03) | 278 (80.12) | |
| III + IV | 76 (21.97) | 69 (19.88) | |
P values are derived from the difference between the discovery data set and the validation data set for clinical characteristics. *, P value <0.05. ECOG, Eastern Cooperative Oncology Group; NA, not available; CR, complete response; SD, stable disease; PD, progressive disease.
Proteomics feature selection and proteomics score construction
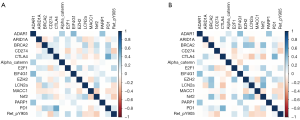
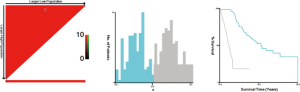
In total, 223 functional proteomics and clinical data were depicted in a heatmap (Figure 1). These proteomics data were reduced to 15 features with the LASSO selection method (Figure 2A). Ten-fold cross validation was used to calculate average decision accuracy with minimum criteria (Figure 2B). Surprisingly, three immune-checkpoint proteins were identified. Based on the selected proteins, including adenosine deaminases acting on RNA 1 (ADAR1), CD274, cytotoxic T-lymphocyte associated protein 4 (CTLA4), programmed death 1 (PD1), and Ret_pY905, a proteomics score was built using the Cox regression model, and all coefficients were presented (Table 2). We further revealed the correlations of 15 proteomics features. There were significant associations with several proteins in the discovery (Figure 3A) and validation sets (Figure 3B). ADAR1 and α-catenin were significantly related in the two cohorts (r=0.294 and 0.160; P<0.001 and 0.002, respectively; Figure S1). We found there were significant correlations in protein levels between PD1 and CTLA4 in the two cohorts (r=0.481 and 0.442; P<0.001 and 0.001, respectively; Figure S1). CD274 (PDL1) and Ret_pY905 were also negatively correlated in the two cohorts (r=0.220 and 0.229; P<0.001 and 0.001, respectively; Figure S1).
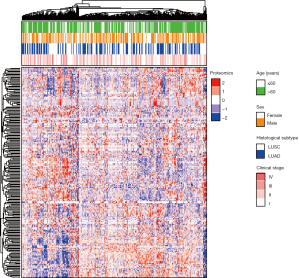
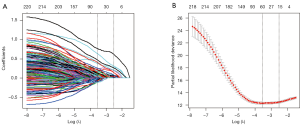
Table 2
| Proteomics features | Coefficients | HR | CI | SE | Z value | P value |
|---|---|---|---|---|---|---|
| ADAR1 | 1.036 | 2.817 | 1.760–4.508 | 0.240 | 4.317 | <0.001* |
| ARID1A | 0.434 | 1.544 | 1.151–2.069 | 0.150 | 2.903 | 0.003* |
| BRCA2 | 0.388 | 1.474 | 1.049–2.073 | 0.174 | 2.232 | 0.025* |
| CD274 | 0.203 | 1.225 | 0.913–1.643 | 0.150 | 1.351 | 0.176 |
| CTLA4 | 0.256 | 1.292 | 0.961–1.738 | 0.151 | 1.696 | 0.089 |
| E2F1 | 0.110 | 1.116 | 0.809–1.540 | 0.164 | 0.670 | 0.502 |
| EZH2 | 0.131 | 1.140 | 0.827–1.570 | 0.163 | 0.799 | 0.424 |
| LCN2a | 0.390 | 1.477 | 1.086–2.009 | 0.157 | 2.485 | 0.012* |
| MACC1 | 0.477 | 1.612 | 1.215–2.138 | 0.144 | 3.308 | <0.001* |
| Nrf2 | 0.233 | 1.262 | 0.925–1.723 | 0.159 | 1.467 | 0.142 |
| PARP1 | 0.567 | 1.763 | 1.157–2.687 | 0.215 | 2.638 | 0.008* |
| PD1 | 0.051 | 1.053 | 0.758–1.463 | 0.168 | 0.307 | 0.758 |
| Ret_pY905 | 0.395 | 1.485 | 1.109–1.988 | 0.149 | 2.654 | 0.007* |
| α-catenin | 0.517 | 1.678 | 1.154–2.438 | 0.191 | 2.711 | 0.006* |
| EIF4G1 | 0.531 | 1.701 | 1.054–2.745 | 0.244 | 2.174 | 0.029* |
*, P value <0.05. HR, hazard ratio; CI, confidence interval; SE, standard errors of coefficients; z value, Wald z-statistic value.
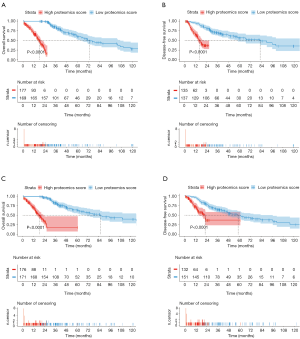
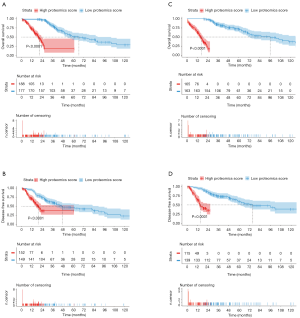
The optimal cut-off value for the proteomics score was 0 using X-tile software in the OS analysis (Figure S2). In the discovery cohort, patients with a high proteomics score showed poorer OS and DFS than those with a low proteomics score [hazard ratio (HR): 5.246; 95% confidence interval (CI): 3.519–7.822; P<0.0001 and HR: 3.470; 95% CI: 2.252–5.346; P<0.0001, respectively; Figure 4A,B). Furthermore, patients with a high proteomics score also showed poorer OS and DFS than those with a low proteomics score (HR: 4.803; 95% CI: 3.166–7.286; P<0.0001 and HR: 2.600; 95% CI: 1.644–4.111; P<0.0001, respectively; Figure 4C,D) in the validation cohort. The results of the sub-analysis of OS and DFS in LUAD and LUSC cohorts are shown in Figure S3.
Development and validation of the proteomics nomogram model
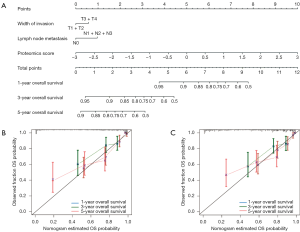
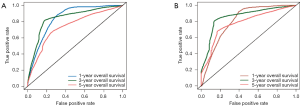
Univariate analysis indicated that the Eastern Cooperative Oncology Group (ECOG) score and outcomes of the first treatment were significantly associated with OS in the discovery set (P=0.027 and 0.026; Table 3). Additionally, depth of invasion, lymph node metastasis, and clinical stage were also significantly related to OS in the discovery set (P=0.001, 0.001, and 0.001; Table 3). The multivariate Cox regression analysis showed that the proteomics score, depth of invasion, and lymph node metastasis were independent predictors of prognosis (P<0.001, 0.044, and 0.001; Table 4). Therefore, this nomogram model was developed and visualized (Figure 5A). Notably, this novel model showed favorable C-indexes of 0.797 (95% CI: 0.765–0.829) and 0.782 (95% CI: 0.747–0.817) for the discovery and validation cohorts, respectively (Table 4). There was a good agreement between the nomogram-estimated probability and actual OS status, as shown in the calibration curves (Figure 5B,C). We used time-dependent ROC curves to estimate the high predictive accuracy of 1, 3, and 5 years in the discovery (AUC =0.844, 0.843, and 0.740; Figure 6A) and validation (AUC =0.808, 0.858 and 0.780; Figure 6B) sets.
Table 3
| Variable | Discovery set (n=346) | |
|---|---|---|
| HR (95% CI) | P value | |
| Sex (male versus female) | 0.967 (0.695–1.345) | 0.838 |
| Age (years) (>60 versus ≤60) | 1.234 (0.860–1.768) | 0.270 |
| Smoking indicator (≤3 versus >3) | 0.932 (0.675–1.288) | 0.672 |
| ECOG score (≤2 versus >2) | 0.294 (0.037–2.323) | 0.027* |
| History of other malignancy (yes versus no) | 1.043 (0.639–1.701) | 0.863 |
| Histological subtype (LUAD versus LSCC) | 1.024 (0.742–1.414) | 0.884 |
| Outcome of first treatment (CR versus SD + PD) | 0.405 (0.129–1.274) | 0.026* |
| Depth of invasion (T1 + T2 versus T3 + T4) | 0.538 (0.330–0.876) | 0.001* |
| Lymph node metastasis (N0 versus N1 + N2 + N3) | 0.549 (0.389–0.775) | 0.001* |
| Distant metastasis (M0 versus M1a + M1b) | 0.454 (0.124–1.665) | 0.125 |
| Clinical stage (I + II versus III + IV) | 0.529 (0.345–0.809) | 0.001* |
| Proteomics score (high versus low) | 5.246 (3.519–7.822) | <0.001* |
P value is derived from univariate analysis of overall survival based on the discovery set. *, P value <0.05. ECOG, Eastern Cooperative Oncology Group; NA, not available; CR, complete response; SD, stable disease; PD, progressive disease.
Table 4
| Intercept and variable | Model | ||
|---|---|---|---|
| B | Hazard ratio (95% CI) | P value | |
| Proteomics score | 0.914 | 2.494 (2.098–2.965) | <0.001* |
| Depth of invasion | 0.308 | 1.461 (1.076–2.042) | 0.044* |
| Lymph node metastasis | 0.521 | 1.684 (1.211–2.342) | 0.001* |
| C-index | |||
| Training cohort | 0.797 (0.765–0.829) | ||
| Validation cohort | 0.782 (0.747–0.817) | ||
P values were obtained from the multivariate regression analysis between the overall survival and each clinical factor. *, P value <0.05. NSCLC, non-small cell lung cancer.
Bioinformation analysis of genes based on selection proteomics
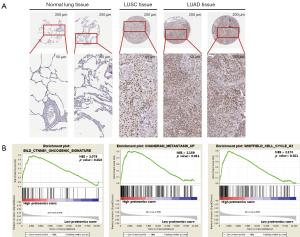
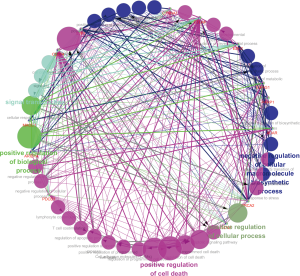
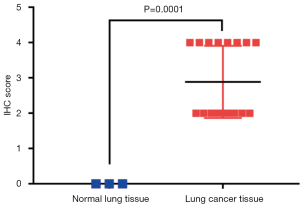
We further evaluated selection genomics as a component of proteomics score. ADAR1, CD274, CTLA4, BRCA2, and PD1 were significantly associated with negative regulation of developmental processes, immune system processes, and cell surface receptor signaling pathways [false discovery rate (FDR): 0.001, 0.006, and 0.015; Table 5]. In GO analysis, we also found that CD274, CTLA4, and PD1 were related to cell adhesion molecule (CAM) pathways (FDR: 0.014). Moreover, BRCA2, α-catenin, E2F transcription factor 1 (E2F1), and RET were associated with cancer pathways (FDR: 0.014). ADAR1 was the protein with the highest HR and shown significantly increased expression (LUAD/LUSC tissue versus normal lung tissue; P=0.001; Figure 7A and Figure S4) in TMA analysis of the HPAS. GSEA revealed that ADAR1 was correlated with oncogenic signature, metastasis, and cell-cycle G2 phase (NES =2.079, 2.159, and 2.174; P=0.013, 0.011, and 0.011; Figure 7B). The association between PPIs and GO/KEGG results were confirmed and visualized using ClueGO (Figure 8). Consequently, in the complex network, the 15 selected genomics were mostly associated with immune processes, cancer signaling pathways, and several important biological processes.
Table 5
| GO term: biological process | FDR | Matching proteins in network (labels) |
|---|---|---|
| Negative regulation of developmental process | 0.001* | BRCA2, CTLA4, α-catenin, E2F1, EZH2, NFE2L2, PD1 |
| Response to oxygen-containing compound | 0.001* | BRCA2, α-catenin, E2F1, EIF4G1, EZH2, NFE2L2, PARP1, RET |
| Negative regulation of multicellular organismal process | 0.002* | BRCA2, CD274, CTLA4, α-catenin, EZH2, NFE2L2, PD1 |
| Negative regulation of immune system process | 0.006* | ADAR1, CD274, CTLA4, NFE2L2, PD1 |
| Cell surface receptor signaling pathway | 0.015* | ADAR1, CD274, CTLA4, α-catenin, E2F1, EIF4G1, PARP1, RET |
| T cell costimulation | 0.015* | CD274, CTLA4, PD1 |
| Multicellular organismal development | 0.020* | ADAR1, ARID1A, BRCA2, α-catenin, E2F1, EIF4G1, EZH2, PARP1, PD1, RET |
| KEGG: pathway description | Matching proteins in network (IDs) | |
| Cell adhesion molecules (CAMs) | 0.014* | CD274, CTLA4, PD1 |
| Pathways in cancer | 0.014* | BRCA2, α-catenin, E2F1, RET |
*, P value <0.05. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas.
Discussion
To the best of our knowledge, this is the first study predicting the prognosis of patients with lung cancer using a proteomics model generated from LUAD and LUSC cohorts from TCGA dataset. Based on functional proteomics analysis of samples from these patients, we developed a proteomics score and nomogram and validated this model as a tool for individualized prediction of OS. This novel risk stratification model using machine learning not only predicted prognosis but also improved risk-adapted treatment in NSCLC.
Precise evaluation of prognosis using mRNA expression data remains challenging in the clinical setting. Currently, different staging systems combining image features and clinical risk factors may also perform well (25-29). However, their levels of accuracy and robustness are unsatisfactory (30,31). We used a proteomics analysis model to transform RPPA data into low-dimensional proteomics features, which were used to estimate patient prognosis. All 223 features from NSCLC proteomics were reduced to 15 proteomics features using the LASSO algorithm and chosen via 10-fold cross-validation to build a proteomics signature. Patients with high proteomics scores showed significantly poorer OS or DFS than those with low proteomics scores. Previous studies also showed that proteomics could be used to predict the prognosis of different patients with cancer (20,32,33). In contrast to previous studies, our study indicated that this novel model of predicting prognosis via combining proteomics may help clinicians to accurately predict the prognosis of patients with NSCLC who underwent partial pneumonectomy.
In our study, we found that there were significant correlations among 15 proteins in both the discovery and validation sets. Similar to previous reports, CTLA4 was positively related to PD1 at the protein level, and combined treatment with CTLA4 and PD1 blocking immunotherapy has been reported in metastatic melanoma, indicating that this therapy may also be suitable for the treatment of NSCLC (34-36). Additionally, we showed that CD274 (PDL1) was negatively associated with Ret_pY905 protein in the two cohorts. In addition to epidermal growth factor receptor mutations and ALK fusions, driver fusions involving RET and ROS1 as well as mutations in KRAS, human epidermal growth factor receptor 2, and BRAF have also been identified in LUAD. Interestingly, our results suggested that immune checkpoint blockade (ICB) therapy may be effective for driver-negative cases (37-39).
In addition, previous studies have reported that several clinical risk factors, such as depth of invasion (tumor size), are related to poor prognosis in NSCLC cases (40-42). Similar to previous studies, univariate analysis of OS in the discovery set showed that most clinical risk factors (e.g., ECOG score, outcome of first treatment, depth of invasion, lymph node metastasis, and clinical stage) were significantly associated with prognosis. According to multivariate analysis of variance, depth of invasion, lymph node metastasis, and proteomics score were independent risk factors of poor OS in the discovery cohort. Considering the above factors, we developed a proteomics nomogram that incorporated the proteomics score, depth of invasion, and lymph node metastasis. The nomogram could be a tool for developing individualized treatment strategies. To the best of our knowledge, the use of proteomics scores and proteomics nomograms for OS prediction has not been previously reported. The proteomics model showed favorable consistency in the discovery cohort, and the outcome was verified in the validation cohort. Time-dependent curves based on the proteomics nomograms in the two cohorts demonstrated that using the proteomics nomogram could precisely evaluate the OS for 1, 3, and 5 years in the discovery and validation sets.
Bioinformatics analysis identified 15 genes that were enriched in negative regulation of developmental processes, immune system processes, cell surface receptor signaling pathways, and other important processes. CD274, CTLA4, and PD1 were associated with CAMs, and BRCA2, α-catenin, E2F1, and RET were related to pathways in cancer. These results supported the involvement in important mechanisms facilitating tumor formation. We validated the ADAR1 protein level based on the TMA from the HPAS. ADAR1 showed significantly higher expression in LUAD and LUSC tissues than in normal lung tissues. Moreover, a previous study reported that the RNA-editing protein, ADAR1, was related to tumor recurrence, invasiveness, and migration of LUAD cells (43,44). Using GSEA, we also discovered that ADAR1 was significantly correlated with the CTNNB1 oncogenic signature, metastasis, and cell-cycle G2 phase, suggesting potential therapeutic applications in LUAD. This PPI indicated that proteomics may play an important role in the regulation of tumorigenesis and in treatment decisions in patients with lung cancer.
Our study had two limitations. First, all NSCLC samples were collected from TCGA and were not validated by multicenter cohorts. Our model may perform differently for data collected from other centers. Thus, much larger datasets must be collected from multiple centers, and the robustness and reproducibility of our proposed proteomics model needs to be investigated. Second, we did not examine whether these proteins played key roles in NSCLC via molecular analyses. Thus, future studies are needed to identify the mechanisms through which these proteins are involved in the pathogenesis of NSCLC.
In conclusion, we demonstrated that the proteomics score and proteomics nomograms may be used to predict prognosis in patients with NSCLC after surgery using TCGA dataset. According to GSEA, GO analysis, and KEGG pathway analysis, the selected proteins were enriched in cancer pathways and immune escape. These findings provided novel insights into which patients with NSCLC may benefit most from partial pneumonectomy, particularly with regard to future clinical trials of targeted treatments or ICB treatments combined with surgery.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.39). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent and ethical approval were obtained. This retrospective study was approved by our institutional review board and Ethical Committee (NFEC-201208-K3).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7:12474. [Crossref] [PubMed]
- Wood DE, Kazerooni EA, Baum SL, et al. Lung Cancer Screening, Version 3.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2018;16:412-41. [Crossref] [PubMed]
- Wakelee H, Kelly K, Edelman MJ. 50 Years of Progress in the Systemic Therapy of Non–Small Cell Lung Cancer. Am Soc Clin Oncol Educ Book 2014;177-89. [Crossref] [PubMed]
- Lang-Lazdunski L. Surgery for nonsmall cell lung cancer. Eur Respir Rev 2013;22:382-404. [Crossref] [PubMed]
- Engelhardt KE, Feinglass JM, DeCamp MM, et al. Treatment trends in early-stage lung cancer in the United States, 2004 to 2013: A time-trend analysis of the National Cancer Data Base. J Thorac Cardiovasc Surg 2018;156:1233-46.e1. [Crossref] [PubMed]
- Tang H, Wang S, Xiao G, et al. Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies. Ann Oncol 2017;28:733-40. [Crossref] [PubMed]
- Yin Y, Zhang Q, Zhang H, et al. Molecular Signature to Risk-Stratify Prostate Cancer of Intermediate Risk. Clin Cancer Res 2017;23:6-8. [Crossref] [PubMed]
- Ye IC, Fertig EJ, DiGiacomo JW, et al. Molecular Portrait of Hypoxia in Breast Cancer: A Prognostic Signature and Novel HIF-Regulated Genes. Mol Cancer Res 2018;16:1889-901. [Crossref] [PubMed]
- Yang L, Taylor J, Eustace A, et al. A Gene Signature for Selecting Benefit from Hypoxia Modification of Radiotherapy for High-Risk Bladder Cancer Patients. Clin Cancer Res 2017;23:4761-8. [Crossref] [PubMed]
- Yang L, Roberts D, Takhar M, et al. Development and Validation of a 28-gene Hypoxia-related Prognostic Signature for Localized Prostate Cancer. EBioMedicine EBioMedicine 2018;31:182-9. [Crossref] [PubMed]
- Kunz M, Löffler-Wirth H, Dannemann M, et al. RNA-seq analysis identifies different transcriptomic types and developmental trajectories of primary melanomas. Oncogene 2018;37:6136-51. [Crossref] [PubMed]
- Haldrup C, Mundbjerg K, Vestergaard EM, et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J Clin Oncol 2013;31:3250-8. [Crossref] [PubMed]
- Shukla S, Evans JR, Malik R, et al. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. J Natl Cancer Inst 2016;109: [Crossref] [PubMed]
- Nagy Á, Pongor LS, Szabó A, et al. KRAS driven expression signature has prognostic power superior to mutation status in non-small cell lung cancer. Int J Cancer 2017;140:930-7. [Crossref] [PubMed]
- Li YY, Yang C, Zhou P, et al. Genome-scale analysis to identify prognostic markers and predict the survival of lung adenocarcinoma. J Cell Biochem 2018;119:8909-21. [Crossref] [PubMed]
- Li L, Feng T, Qu J, et al. LncRNA Expression Signature in Prediction of the Prognosis of Lung Adenocarcinoma. Genet Test Mol Biomarkers 2018;22:20-8. [Crossref] [PubMed]
- Li B, Cui Y, Diehn M, et al. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non-Small Cell Lung Cancer. JAMA Oncol 2017;3:1529-37. [Crossref] [PubMed]
- Spurrier B, Ramalingam S, Nishizuka S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc 2008;3:1796-808. [Crossref] [PubMed]
- Gulmann C, Sheehan KM, Kay EW, et al. Array-based proteomics: mapping of protein circuitries for diagnostics, prognostics, and therapy guidance in cancer. J Pathol 2006;208:595-606. [Crossref] [PubMed]
- Nishizuka S, Spurrier B, Honkanen P, et al. Application of quantitative proteomic analysis for cancer therapy using "reverse-phase" protein lysate microarrays. Gan To Kagaku Ryoho 2008;35:200-5. [PubMed]
- Belczacka I, Latosinska A, Metzger J, et al. Proteomics biomarkers for solid tumors: Current status and future prospects. Mass Spectrom Rev 2019;38:49-78. [Crossref] [PubMed]
- Swiatly A, Horala A, Matysiak J, et al. Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery. Int J Mol Sci 2018; [Crossref] [PubMed]
- Iglesias-Gato D, Thysell E, Tyanova S, et al. The Proteome of Prostate Cancer Bone Metastasis Reveals Heterogeneity with Prognostic Implications. Clin Cancer Res 2018;24:5433-44. [Crossref] [PubMed]
- Wu Y, Xu L, Yang P, et al. Survival Prediction in High-grade Osteosarcoma Using Radiomics of Diagnostic Computed Tomography. EBioMedicine 2018;34:27-34. [Crossref] [PubMed]
- Lee J, Li B, Cui Y, et al. A Quantitative CT Imaging Signature Predicts Survival and Complements Established Prognosticators in Stage I Non-Small Cell Lung Cancer. Int J Radiat Oncol Biol Phys 2018;102:1098-106. [Crossref] [PubMed]
- Sollini M, Cozzi L, Chiti A, et al. Texture analysis and machine learning to characterize suspected thyroid nodules and differentiated thyroid cancer: Where do we stand? Eur J Radiol 2018;99:1-8. [Crossref] [PubMed]
- Wu S, Zheng J, Li Y, et al. Development and Validation of an MRI-Based Radiomics Signature for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. EBioMedicine 2018;34:76-84. [Crossref] [PubMed]
- Chen S, Zhu Y, Liu Z, et al. Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol 2017;90:198-204. [Crossref] [PubMed]
- Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. [Crossref] [PubMed]
- Berenguer R, Pastor-Juan MD, Canales-Vázquez J, et al. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology 2018;288:407-15. [Crossref] [PubMed]
- Schwartz GW, Petrovic J, Zhou Y, et al. Differential Integration of Transcriptome and Proteome Identifies Pan-Cancer Prognostic Biomarkers. Front Genet 2018;9:205. [Crossref] [PubMed]
- Krisp C, Parker R, Pascovici D, et al. Proteomic phenotyping of metastatic melanoma reveals putative signatures of MEK inhibitor response and prognosis. Br J Cancer 2018;119:713-23. [Crossref] [PubMed]
- Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015;14:561-84. [Crossref] [PubMed]
- Hellmann MD, Rizvi NA, Goldman JW, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): results of an open-label, phase 1, multicohort study. Lancet Oncol 2017;18:31-41. [Crossref] [PubMed]
- Chae YK, Arya A, Iams W, et al. Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC). J Immunother Cancer 2018;6:39. [Crossref] [PubMed]
- Kazandjian D, Suzman DL, Blumenthal G, et al. FDA Approval Summary: Nivolumab for the Treatment of Metastatic Non-Small Cell Lung Cancer With Progression On or After Platinum-Based Chemotherapy. Oncologist 2016;21:634-42. [Crossref] [PubMed]
- Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res 2016;22:4585-93. [Crossref] [PubMed]
- Dong ZY, Zhang JT, Liu SY, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 2017;6:e1356145. [Crossref] [PubMed]
- Ye T, Deng L, Xiang J, et al. Predictors of Pathologic Tumor Invasion and Prognosis for Ground Glass Opacity Featured Lung Adenocarcinoma. Ann Thorac Surg 2018;106:1682-90. [Crossref] [PubMed]
- Tsutani Y, Miyata Y, Nakayama H, et al. Oncologic outcomes of segmentectomy compared with lobectomy for clinical stage IA lung adenocarcinoma: propensity score-matched analysis in a multicenter study. J Thorac Cardiovasc Surg 2013;146:358-64. [Crossref] [PubMed]
- Li S, Zhu R, Li D, et al. Prognostic factors of oligometastatic non-small cell lung cancer: a meta-analysis. J Thorac Dis 2018;10:3701-13. [Crossref] [PubMed]
- Wang C, Zou J, Ma X, et al. Mechanisms and implications of ADAR-mediated RNA editing in cancer. Cancer Lett 2017;411:27-34. [Crossref] [PubMed]
- Amin EM, Liu Y, Deng S, et al. The RNA-editing enzyme ADAR promotes lung adenocarcinoma migration and invasion by stabilizing FAK. Sci Signal 2017; [Crossref] [PubMed]


