
LncRNA SNHG1 upregulates ROCK2 to reduce cisplatin sensitivity of NSCLC cells by targeting miR-101-3p
Introduction
Lung cancer results in high mortality worldwide, while non-small cell lung cancer (NSCLC) accounts for more than 85% of newly diagnosed lung cancer cases (1,2). Chemotherapy is the most important method for patients with advanced lung cancer, and cisplatin, oxaliplatin and carboplatin are the most commonly used chemotherapy drugs in clinical practice. Cytotoxicity of these drugs is mainly caused by the formation of platinum-DNA complexes that induce DNA damage and cell apoptosis (3). The rate of cisplatin chemotherapy is only 30–40%, and the resistance of tumor cells to chemotherapy drugs is the main factor that affects the efficacy of chemotherapy (4,5). Therefore, exploration of the molecular mechanism of cisplatin resistance in NSCLC cells is of great significance in improving the survival rate and prognosis of lung cancer patients.
Long non-coding RNAs (lncRNAs) play an important role in the development of various cancers, and they have become a new hot spot in tumor research due to their significant tumor tissue specificity and potential applications in tumor diagnosis and prognosis prediction (6,7). LncRNAs are RNAs with a length of over 200 bp, with either no or a small opening reading frame, that cannot or rarely encodes a protein. They are usually composed of multiple spliced exons, are transcribed by RNA polymerase II, and contain a histone modification similar to that of encoded proteins (8,9). In recent years, an increasing number of lncRNAs have been found to be closely related with chemotherapy drug resistance (10,11) and studies have found that lncRNAs can be used as targets of chemotherapy drug resistance (12,13).
LncRNA SNHG1 is found to be abnormally expressed in breast cancer (14), nasopharyngeal carcinoma (15), colon cancer (16) and a variety of other malignant tumor tissues, and plays a very important role in the regulation of tumor cell proliferation, migration and resistance to chemotherapy drugs (17,18). Previous research has found that upregulated lncRNA SNHG1 contributes to the progression of NSCLC, but the effect of SNHG1 on cisplatin sensitivity of NSCLC cells is unclear (19). In this study, we compared the expression of SNHG1 between cisplatin-sensitive and insensitive NSCLC tissues and explored the molecular mechanism used by SNHG1 to regulate the sensitivity of NSCLC cells to cisplatin by analyzing the expression of other molecules, such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), U6 and miR-101-3p, in vitro. We found that SNHG1 is highly expressed in cisplatin-insensitive NSCLC tissues, and that it can promote the expression of ROCK2 by inhibiting the expression of miR-101-3p, thereby reducing the sensitivity of NSCLC cells to cisplatin in vitro.
Methods
Patients and tissues
Sixty-four NSCLC patients were diagnosed and treated through surgery at Nanjing Jinling Hospital, Nanjing University School of Medicine from September 2016 to September 2017. A total of 64 patients were operated on for NSCLC, and all patients were included in this study. Fresh tumor tissue from surgical resection was prepared as a single cell suspension, followed by a chemotherapeutic drug susceptibility test. Based on the drug susceptibility test results, the 64 NSCLC patients were divided into two groups: the cisplatin sensitive group (sensitive, n=32), and the cisplatin insensitive group (insensitive, n=32).
Additionally, neither of the NSCLC patients had other malignant tumors, chronic cardiovascular and cerebrovascular diseases, infectious diseases, and had not received radiotherapy, chemotherapy or immunotherapy before surgery. Informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Nanjing Jinling Hospital, Nanjing University School of Medicine (ethical approval number: 2016-037).
Cell and cell culture
A549 (CCL-185, ATCC, VA, USA), NCI-H520 (HTB-182, ATCC, VA, USA), A549/DDP (CL-0519, Procell, Wuhan, China) and NCI-H520/DDP (CL-0758, Procell, Wuhan, China) cells were cultured in DMEM medium (12491-15, Thermo Fisher, CA, USA), to which 10% fetal bovine serum (10100-147, Thermo Fisher, CA, USA) and 1% penicillin-streptomycin (15640055, Thermo Fisher, CA, USA) were added. In addition, 1 ug of DDP (H37021357, Qilu Pharmaceutical Co., Ltd., Shandong, China) was also added to the culture medium of the A549/DDP and NCI-H520/DDP cells.
Quantitative real-time polymerase chain reaction (RT-qPCR)
RT-qPCR was used to detect the expression of GAPDH mRNA, U6, miR-101-3p and lncRNA SNHG1 in tissues and cells. TRIzol was used to extract total RNA of the tissues or cells. For tissues, liquid nitrogen and TRIzol were simultaneously added to the mortar, and the tissues were ground into a powder. For the cells, TRIzol was added directly to the cells, after blowing several times back and forth. The extracted RNA was reverse transcribed into cDNA using the PrimeScript™ RT Master Mix reverse transcription kit (RR036B, Takara, Beijing, China). PCR parameters used were as follows: 37 °C for 60 minutes and 85 °C for 5 seconds. Twenty µL of the fluorescence RT-qPCR system mixture was prepared according to the SYBR Green qPCR Master Mix kit instructions (638320, TakaRa, Beijing, China) and amplified using an ABI 7500 fluorescence quantitative PCR instrument (Applied Biosystems, MD, USA). The PCR parameters were set as follows: 40 cycles of 95 °C for 30 s (90 °C for 5 s, 65 °C for 30 s). The following PCR primers were used: U6-F: 5'-AUAAAUCCCUUUACACCUCTT-3', U6-R: 5'-AAUAAAUCCCUUUACACCUCTT-3'; GAPDH-F: 5'-GATGAACCTAAGCTGGGACCC--3', GAPDH-R: 5'-TGTGAACGGATTTGGCCGTA-3'; miR-101-3p-F: 5'-ACACTCCAGCTGGGTACAGUACUGUGAUA-3', miR-101-3p-R: 5'-TGGTGTCGTGGAGTCG-3'; SNHG1-F: 5'-AGGCTGAAGTTACAGGTC-3', SNHG1-R: 5'-TTGGCTCCCAGTGTCTTA-3'.
Cell transfection
The si-RNA for SNHG1 knockdown (si-SNHG1), control knockdown (si-NC), ROCK2 knockdown (si-ROCK2) and Hsa-miR-101-3p-mimic and Hsa-miR-31-5p-NC were designed and synthesized by Shenggong Bioengineering Co., Ltd. (Shanghai, China). si-RNA, si-NC, microRNA-mimic and microRNA-NC were directly transferred into cells using Lipofectamine™ 2000 transfection reagent (11668019, Invitrogen, CA, USA).
Wild type or mutation mRNA 3'-UTR of SNHG1 and 3'-UTR of ROCK2 were first connected to pisCHECK2 (Promega, WI, USA) and then transfected into cells as si-RNAs. LV-SNHG1/LV-ROCK2 is a plasmid that was used to upregulate the expression of SNHG1/ROCK2, and it was constructed as follows: the full-length sequence of the SNHG1/ROCK2 molecule was synthesized using PCR at Shenggong Bioengineering Co., Ltd. (Shanghai, China), and it was inserted into pcDNA3.1 (+) (Invitrogen, CA, USA) plasmid via the KpnI and EcoRI sites, and was then transfected into cells as a si-RNA. The control upregulator (LV-NC) was transfected only into the pcDNA3.1 (+) plasmid.
Cell counting kit-8 assay (CCK8)
CCK8 kit was used to measure cisplatin cytotoxicity. In brief, 5×103 cells/well were seeded into 96 wells, and 0, 1, 2, 5, 10, 20 or 30 µM of cisplatin was added for 24 hours of culture. Then, all the wells were incubated with the CCK8 reagent for 4 h, and the optical density at 450 nm (OD450) was used to show cell viability. Moreover, in order to measure the concentration of cisplatin that accounted for a half maximal inhibitory concentration (IC50), A549/DDP and NCI-H520/DDP cells required 40, 50, 60, 100 µM of cisplatin for the drug toxicity test.
Western blotting
The tissue or cell lysates were separated using SDS-PAGE and then transferred to PVDF membranes. The primary antibody selected was anti-ROCK2 (ab71598; 1:1,000) or anti-GAPDH (ab9484; 1:3,000), while the secondary antibody selected was goat anti-rabbit (ab150077; 1:1,000). The samples were incubated with the primary antibody overnight at 4 °C and with the secondary antibody for 1 hour at room temperature.
Statistical analysis
Data are presented as mean ± standard deviation and all analyses were performed using SPSS 20.0 software. Student’s t-test or chi-square test were used to compare differences between two groups. Kaplan-Meier analysis was used to generate survival plots and the significance of the differences was assessed using the log-rank test. Pearson correlation was used to analyze the correlation between lncRNA SNHG1 and miR-101-3p in 64 NSCLC tumor tissues. A P value of <0.05 was considered to indicate statistical significance.
Results
SNHG1 is upregulated in cisplatin insensitive NSCLC tissues and cells
RT-qPCR was used to detect the expression of lncRNA SNHG1 in 32 samples of cisplatin sensitive NSCLC tissues and 32 samples of cisplatin insensitive NSCLC tissues. The results show that the expression of SNHG1 in cisplatin insensitive NSCLC tissues was significantly higher than that of cisplatin sensitive tissues (Figure 1A). Moreover, the expression of SNHG1 in DPP resistant NSCLC cells was found to be significantly higher than that in wild type NSCLC cells (Figure 1B).
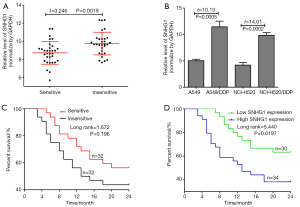
We analyzed the correlation between cisplatin sensitivity and clinicopathological features of NSLCL patients. As shown in Table 1, the expression of SNHG1 was not associated with age, gender, tumor-node-metastasis (TNM) stage, T stage or N stage. The 2-year survival of patients with cisplatin sensitivity was higher than that of cisplatin insensitive patients, but the difference was not significant (log rank=1.672, P=0.196) (Figure 1C). In addition, the 64 NSCLC patients were divided into two groups based on the ratio of the expression of lncRNA SNHG1 in cancer tissues: 30 patients were accordingly placed in the low expression of SNHG1 group (ratio < median value of the 64 NSCLC patients) and 34 patients were placed in the high expression of SNHG1 group (ratio ≥ median value of the 64 NSCLC patients). The 2-year survival of patients with low expression of SNHG1 was found to be significantly higher than that of patients with high expression of SNHG1 (log rank=5.440, P=0.0197) (Figure 1D).
Table 1
| Variables | Case | Cisplatin | χ2 | P | |
|---|---|---|---|---|---|
| Sensitive | Insensitive | ||||
| Gender | 0.277 | 0.599 | |||
| Male | 42 | 20 | 22 | ||
| Female | 22 | 12 | 10 | ||
| Age, year | 0.259 | 0.611 | |||
| <60 | 38 | 18 | 20 | ||
| ≥60 | 26 | 14 | 12 | ||
| TNM stage | 3.373 | 0.185 | |||
| I | 25 | 16 | 9 | ||
| II | 18 | 8 | 10 | ||
| III | 21 | 8 | 13 | ||
| T | 2.250 | 0.134 | |||
| T1–T2 | 32 | 19 | 13 | ||
| T3–T4 | 32 | 13 | 19 | ||
| N | 3.216 | 0.073 | |||
| N0 | 25 | 16 | 9 | ||
| N1–N3 | 39 | 16 | 23 | ||
NSCLC, non-small cell lung cancer; TNM, tumor-node-metastasis.
SNHG1 regulates cisplatin sensitivity of NSCLC cells
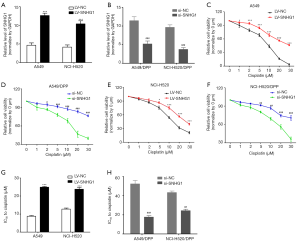
We upregulated the expression of SNHG1 in A549 and NCI-H520 cells by transfecting LV-SNHG1 into the cells (Figure 2A), and downregulated the expression of SNHG1 in A549/DDP and NCI-H520/DDP cells by transfecting si-SNHG1 into the cells (Figure 2B). As shown in Figure 2C,D,E,F,G,H, CCK8 viability assay shows that SNHG1 upregulation in A549 and NCI-H520 cells resulted in increased viability upon cisplatin treatment and lower IC50 values for cisplatin, while SNHG1 downregulation resulted in decreased viability.
SNHG1 binds to miR-101-3p in NSCLC cells
We measured the expression of miR-101-3p in NSCLC tissues and NSCLC cells, and found that the expression of miR-101-3p in cisplatin sensitive NSCLC tissues and cells is significantly higher than that of cisplatin insensitive cells, while a negative correlation between SNHG1 and miR-101-3p were found in NSCLC tissues (Figure 3A,B,C).
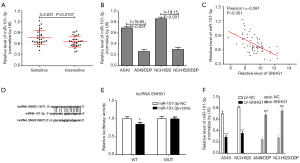
We searched for binding sites between miR-101-3p and SNHG1 using the starbase database (Figure 3D). Luciferase gene reporter system was used to confirm that miR-101-3p can regulate SNHG1 expression by binding to the SNHG1 3'-UTR end. The results show that (Figure 3E) transfection with a miR-101-3p-mimic significantly decreased WT type 3'-UTR luciferase activity (P<0.001) in A549 cells. However, the same was not observed in the mutated type (MUT). Further, as shown in Figure 3F, lncRNA SNHG1 downregulation could increase the expression of miR-101-3p, and its upregulation could decrease the same. The above results suggest that miR-101-3p and lncRNA SNHG1 can inhibit the expression of the other in NSCLC cells.
SNHG1/miR-101-3p/ROCK2 axis mediates cisplatin resistance in NSCLC cells
Previous research has shown that downregulation of miR-101 contributes to epithelial-mesenchymal transition in cisplatin resistance of NSCLC cells by targeting ROCK2. ROCK2, the Rho-associated Frizzled protein kinase 2, is a key effector 236 downstream of the Rho family. In this study, we also measured the expression of ROCK2 protein, and found that the expression of ROCK2 protein in cisplatin sensitive NSCLC tissues is significantly lower than that of cisplatin insensitive tissues (Figure 4A), with a negative correlation with miR-101-3p in NSCLC tissues (Figure 4B).
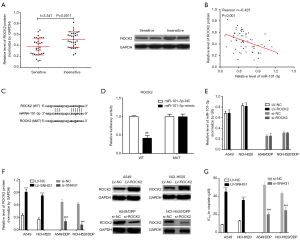
We also searched for binding sites between miR-101-3p and ROCK2 using the starbase database (Figure 4C). Luciferase gene reporter system was used to confirm that miR-101-3p can regulate ROCK2 expression by binding to the ROCK2 3'-UTR end. The results show that (Figure 4D) transfection with the miR-101-3p-mimic significantly decreases WT type 3'-UTR luciferase activity in A549, but it did not work in MUT. However, neither ROCK2 downregulation or upregulation could change the expression of miR-101-3p in NSCLC cells (Figure 4E). These results suggest that ROCK2 is a target gene of miR-101-3p. In addition, CCK8 viability assay showed that ROCK2 upregulation in A549 and NCI-H520 cells results in increased viability upon cisplatin treatment and lower IC50 values for cisplatin, while ROCK2 downregulation results in the decrease of the same (Figure 4F,G).
Discussion
Since the beginning of the 21st century, the incidence of cancer has been increasing in most countries. Therefore, research on the pathogenesis and treatment of tumors has become a primary focus of modern medicine. At present, along with the gradual introduction of new drugs and the development of medical technology, great progress has been made regarding tumor treatment. Cancer treatment drugs, including chemotherapy and targeted drug therapy, are currently the most widely used form of treatment. However, in clinical applications, tumor cells often develop resistance to drugs, resulting in treatment failure and disease recurrence, which greatly affects the cure rate of cancer patients (20). In recent years, along with the widespread application of second-generation sequencing technology, researchers have found that non-protein-encoding gene sequences are closely related to the occurrence and development of cancer and drug tolerance (21,22). Among them, lncRNA is a hotspot in tumor resistance mechanism research. Many reports indicate that lncRNAs are involved in the development of drug-resistant phenotypes of tumors (23).
In this paper, we found that SNHG1 is upregulated in cisplatin insensitive NSCLC tissues and cells, and it can regulate cisplatin sensitivity of NSCLC cells in vitro. At present, many studies on lncRNAs in NSCLC cancer have been published, and many lncRNAs, such as DANCR (24), MEG3 (25) and HOTAIR (26), have been identified. lncRNA SNHG1 is located on chromosome 11 and has a full length of 780 bp. Previous studies have confirmed that SNHG1 acts as a proto-oncogene in a variety of malignancies (17). Yun Cui found that upregulated lncRNA SNHG1 contributes to the progression of NSCLC through the inhibition of miR-101-3p and activation of the Wnt/β-catenin signaling pathway (19). lncRNA can prevent tumor cells from apoptosis under the action of drugs by blocking the cell cycle, regulating the apoptosis transduction pathway, mediating protooncogene and tumor suppressor gene expressions, and producing tumor resistance. In combination with the study by Cui et al., lncRNA SNHG1 was found to be likely to affect cisplatin sensitivity of NSCLC cells by regulating the expression of miR-101-3p.
Fortunately, our study found that miR-101-3p is downregulated in cisplatin insensitive NSCLC tissues and cells, and is negatively correlated with SNHG1 expression in NSCLC tissues. At the same time, luciferase gene reporter system was used to confirm that SNHG1 is a target gene of miR-101-3p, which inhibits its expression in NSCLC cells. As is well known, lncRNAs and miRNAs are non-coding RNAs that exert their biological functions by regulating the RNA that encodes the protein (27,28). Luciferase gene reporter system was also used to confirm that ROCK2 is a target gene of miR-101-3p, and that miR-101-3p can target the inhibition of ROCK2 expression in NSCLC cells. More importantly, ROCK2 can also regulate cisplatin sensitivity of NSCLC cells in vitro.
Recent studies have shown that ROCK2 is expressed in malignant tumors, such as breast cancer (29), colon cancer (30) and prostate cancer (31). Previous research has shown that ROCK2 is mainly present in the cytoplasm, regulating cytoskeletal component movement through its action on actin polymerization, myosin contraction, cell adhesion and microtubule dynamics, which change cell morphology, polarity and promote cell proliferation that participates in the invasion and metastasis of malignant tumors (32). ROCK2 has been rarely reported in lung cancer, but Vigil et al. found that ROCK1 and ROCK2 are required for NSCLC anchorage-independent growth and invasion (33). A recent study has indicated that miR-101 targets the inhibition of ROCK2 expression, whereas downregulation of miR-101 expression promotes the resistance of NSCLC cells to cisplatin by upregulating the expression of ROCK2 (34). Therefore, our results provide novel mechanistic insights into the role of SNHG1/miR-101-3p/ROCK2 signaling in cisplatin resistance of NSCLC cells.
Conclusions
lncRNA SNHG1 and ROCK2 protein are upregulated, while miR-101-3p is downregulated, in cisplatin insensitive NSCLC tissues and cells. In vitro, SNHG1 and miR-101-3p inhibit each other, but miR-101-3p targets the inhibition of ROCK2 expression. In summary, lncRNA SNHG1 upregulates ROCK2 in order to decrease cisplatin sensitivity of NSCLC cells by targeting miR-101-3p.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.24). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Nanjing Jinling Hospital, Nanjing University School of Medicine (ethical approval number: 2016-037).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China in 2013: an analysis based on urbanization level. Chin J Cancer Res 2017;29:1-10. [Crossref] [PubMed]
- Fikrova P, Stetina R, Hrnciarik M, et al. DNA crosslinks, DNA damage and repair in peripheral blood lymphocytes of non-small cell lung cancer patients treated with platinum derivatives. Oncol Rep 2014;31:391-6. [Crossref] [PubMed]
- Soria JC, Tan DSW, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 2017;389:917-29. [Crossref] [PubMed]
- Fennell DA, Summers Y, Cadranel J, et al. Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat Rev 2016;44:42-50. [Crossref] [PubMed]
- Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res 2012;22:885-98. [Crossref] [PubMed]
- Tani H, Mizutani R, Salam KA, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res 2012;22:947-56. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Hon CC, Ramilowski JA, Harshbarger J, et al. An atlas of human long non-coding RNAs with accurate 5' ends. Nature 2017;543:199-204. [Crossref] [PubMed]
- Sun W, Zu Y, Fu X, et al. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep 2017;38:3347-54. [PubMed]
- Pan J, Li X, Wu W, et al. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett 2016;382:64-76. [Crossref] [PubMed]
- Ma Y, Zhou G, Li M, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling pathway. Neurochem Int 2018;118:233-41. [Crossref] [PubMed]
- Wang Y, Zhang L, Zheng X, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett 2016;382:137-46. [Crossref] [PubMed]
- Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol 2018;118:24-30. [Crossref] [PubMed]
- Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med 2019;23:2351-61. [Crossref] [PubMed]
- Yang H, Wang S, Kang YJ, et al. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol Rep 2018;40:261-71. [PubMed]
- Sun Y, Wei G, Luo H, et al. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene 2017;36:6774-83. [Crossref] [PubMed]
- Wang Q, Li Q, Zhou P, et al. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother 2017;91:906-11. [Crossref] [PubMed]
- Cui Y, Zhang F, Zhu C, et al. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget 2017;8:17785-94. [PubMed]
- Ledzewicz U, Schättler H. Drug resistance in cancer chemotherapy as an optimal control problem. Discrete and Continuous Dynamical Systems - Series B (DCDS-B) 2006. doi:
10.3934/dcdsb.2006.6.129 . - Fan Y, Shen B, Tan M, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J 2014;281:1750-8. [Crossref] [PubMed]
- Wang J, Li L, Wu K, et al. Knockdown of long noncoding RNA urothelial cancer-associated 1 enhances cisplatin chemosensitivity in tongue squamous cell carcinoma cells. Pharmazie 2016;71:598-602. [PubMed]
- Hu L, Chen J, Zhang F, et al. Aberrant long noncoding RNAs expression profiles affect cisplatin resistance in lung adenocarcinoma. Biomed Res Int 2017;2017:7498151. [Crossref] [PubMed]
- Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett 2017;405:46-55. [Crossref] [PubMed]
- Terashima M, Tange S, Ishimura A, et al. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem 2017;292:82-99. [Crossref] [PubMed]
- Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013;13:464. [Crossref] [PubMed]
- Ender C, Krek A, Friedländer MR, et al. A human snoRNA with microRNA-like functions. Mol Cell 2008;32:519-28. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Lane J, Martin TA, Watkins G, et al. The expression and prognostic value of ROCK I and ROCK II and their role in human breast cancer. Int J Oncol 2008;33:585-93. [PubMed]
- Vishnubhotla R, Sun S, Huq J, et al. ROCK-II mediates colon cancer invasion via regulation of MMP-2 and MMP-13 at the site of invadopodia as revealed by multiphoton imaging. Lab Invest 2007;87:1149-58. [Crossref] [PubMed]
- Kroiss A, Vincent S, Decaussin-Petrucci M, et al. Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene 2015;34:2846-55. [Crossref] [PubMed]
- Itoh K, Yoshioka K, Akedo H, et al. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat Med 1999;5:221-5. [Crossref] [PubMed]
- Vigil D, Kim TY, Plachco A, et al. ROCK1 and ROCK2 are required for non-small cell lung cancer anchorage-independent growth and invasion. Cancer Res 2012;72:5338-47. [Crossref] [PubMed]
- Ye Z, Yin S, Su Z, et al. Downregulation of miR-101 contributes to epithelial-mesenchymal transition in cisplatin resistance of NSCLC cells by targeting ROCK2. Oncotarget 2016;7:37524-35. [Crossref] [PubMed]


