
High expression of TOP2A gene predicted poor prognosis of hepatocellular carcinoma after radical hepatectomy
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy worldwide and one of the leading causes of cancer-related death (1). Nearly 70% of these cancer deaths occur in low and middle-income countries. In recent years, the main treatment methods for HCC include surgical resection, local ablation, transhepatic arterial chemotherapy and embolization (TACE), and molecular targeting (2). However, the therapeutic effects are still not satisfactory. In particular, the recurrence of HCC after surgical resection is a key factor affecting the therapeutic effect, and progress has been slowing for many years (3). Clinical prognostic indexes of primary HCC include tumor size, liver cirrhosis, the grade of tumor differentiation, and invasion of microvessels (4-6). Due to the uncertainty in the development of HCC, the above predictors are still not accurate for predicting the prognosis after surgical resection.
Topoisomerase (DNA) II alpha (TOP2A) is the up-regulated gene of the chromosome bond pathway (7). This gene encodes DNA topoisomerase, which can control and change the topological state of DNA during transcription and replication, take part in chromosome agglutination and separation, and relieve kinking stress. Additionally, the abnormal expression of TOP2A gene is often associated with active cell proliferation (8). In studies of nasopharyngeal carcinoma (9), breast cancer (10), endometrial cancer (11), and adrenal cancer (12), it was found that the high expression of TOP2A suggested a poor prognosis. By searching the www.oncomine.org database, it was discovered that TOP2A was found to be highly expressed in HCC tumor tissues. Therefore, this study aimed to retrospectively analyze the expression of TOP2A in HCC and understand the clinicopathological characteristics of HCC to explore its feasibility as a prognostic indicator.
Methods
Tumor samples
A total of 84 HCC and para-tumor paraffin specimens from January 2011 to December 2012 were collected from patients who had undergone radical resection at the Affiliated Hospital of Nantong University. The patients were aged 35 to 74 years old, with no limitation on gender. All cases were single tumors. No other malignant tumors were observed in all patients, and no portal venous carcinoma thrombus was observed by preoperative imaging and intraoperative macroscopic examination. From March 2018 to August 2018, 15 pairs of fresh liver tumor specimens and para-cancer tissues were taken.
Patients’ clinical data
The clinical features of the patients are shown in the table below (Table 1). Among all the cases, the 72 males and 12 females had an average age of 52.92 years (35–74 years), 77 were hepatitis B surface antigen (HBsAg) positive, 63 had significant cirrhosis, and the average tumor size was 4.99 cm (1–15 cm), among which 50 were less than 5 cm, 24 were greater than or equal to 5 cm, and 10 were greater than or equal to 10 cm. Alpha-fetoprotein (AFP) was greater than or equal to 400 ng/mL in 33 cases and <400 ng/mL in 51 cases. The degree of pathological differentiation was determined by Edmonson classification, including grade I (11 cases), grade II (42 cases), grade III (29 cases), and grade IV (2 cases).
Table 1
| Groups | Patients, n | Percentage of total | TOP2A high expression, n | Percentage of total | χ2 | P value |
|---|---|---|---|---|---|---|
| Tumor size | 5.945 | 0.0148 | ||||
| <5 cm | 50 | 59.52% | 25 | 50.00% | ||
| ≥5 cm | 34 | 40.48% | 26 | 76.47% | ||
| Liver cirrhosis | 12.13 | 0.0005 | ||||
| Absent | 21 | 25.00% | 6 | 28.57% | ||
| Present | 63 | 75.00% | 45 | 71.43% | ||
| AFP | ||||||
| <400 ng/mL | 51 | 60.71% | 31 | 60.78% | 0.0002669 | 0.987 |
| ≥400 ng/mL | 33 | 39.29% | 20 | 60.61% | ||
| Differentiation | 5.748 | 0.0165 | ||||
| Grade I−II | 53 | 63.10% | 27 | 50.94% | ||
| Grade III−IV | 31 | 36.90% | 24 | 77.42% | ||
| HBsAg | 0.04084 | 0.8399 | ||||
| Positive | 77 | 91.67% | 47 | 61.04% | ||
| Negative | 7 | 8.33% | 4 | 57.14% | ||
| Five-year survival | 14.51 | <0.0001 | ||||
| Yes | 37 | 44.05% | 14 | 37.84% | ||
| No | 47 | 55.95% | 37 | 78.72% |
Pathological diagnosis and grading
After hematoxylin and eosin (HE) staining, paraffin sections of all patients were accurately diagnosed by 2 pathologists according to the World Health Organization standards, and tumors were graded according to Edmonson grading standards. When differentiated tumor cells appeared in the same specimen, the higher grade was taken as the final result.
Real-time PCR was used to detect the expression of TOP2A in HCC and para-cancer tissues
RNA preparation proceeded as follows: 15 pairs of HCC tumor tissues and related para-cancer tissues were collected, and total RNA was extracted by Trizol.
The synthesis of cDNA chain 1 proceeded as follows: total RNA was reversely transcribed into cDNA, and Beacon Designer was used to design the real time (RT)-PCR primer. TOP2A-F:5'-CATTGAAGACGCTTCGTTATGG-3', TOP2A-R:5'-CAGAAGAGAGGGCCAGTTGTG-3'; Made β-actin-F:5'-ATAGCACAGCCTGGATAGCAACGTAC-3', β-actin-R:5'- CACCTTCTACAATGAGCTGCGTGTG-3'. The quantitative (Q)-PCR reaction system was as follows: SYBR qPCR Mix 10 µL, qPCR upstream and downstream primers 0.8 µL (10 µmol/L), cDNA 2 µL, 50× ROX reference dye 0.4 µL, deionized water to 20 µL. The reaction system computer detection was as follows: predenaturation at 95 °C for 1 min, 40 cycles. Each PCR cycle contained denaturation at 95 °C for 30 s, annealing at 60 °C for 40 s, and a final elongation at 72 °C for 10 min. All samples were set with 3 parallel multiple holes for 3 times of repeated detection. The TOP2A expression was measured using 2−ΔCT method.
Western blotting assay
According to the method reported in the literature (12), 8 samples of tumor and paracancer tissues were homogenized, and total protein was extracted for protein quantification. The samples were then subjected to gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was incubated with 5% skimmed milk at room temperature for 2 h and then mixed with primary TOP2A antibody at dilution of 1:1,000 for overnight incubation at 4 °C. After washing the membrane, the secondary antibody was added for 1 h incubation at room temperature and finally exposed and developed by X-ray. GAPDH was used as the loading control.
The expression of TOP2A in HCC was detected by immunohistochemistry (IHC)
Tissue chip making
Tissue chips were prepared from the HCC tumor samples (n=84) and surrounding tissues, respectively, fixed by formalin, and embedded by paraffin. The thickness of the slices was 4 µm. IHC streptavidin peroxidase staining was used.
Paraffin sections were dewaxed to water and rinsed with distilled water 3 times, endogenous peroxidase was inactivated by 3% H2O2 for 10 min, and antigen repair was performed with sodium citrate for 20 min by phosphate buffer solution (PBS), followed by 3 PBS rinses of 5 min, and 10% goat serum and incubation for 30 min. They were incubated with rabbit anti-TOP2A antibody (1:200) at 37 °C for 2 h, then rinsed with PBS. The second antibody was added and rested at 37 °C for 15 min incubation and PBS rinsing. Mold avidin horseradish enzyme marker chain working liquid was then added at 37 °C for 15 min incubation and PBS rinsing, then chromogenic with diaminobenzidine (DAB), redyed, dehydrated, made transparent, and sealed with rubber. Non-reactive PBS replaced TOP2A primary antibody in negative and positive control experiments, and the rest of the operations were the same as above.
Two double-blinded pathologists evaluated the results of immunohistochemical staining. According to the positive percentage, the evaluated results were divided into the following four levels: 0 (0%), 1 (1–33%), 2 (34–66%), and 3 (67–100%). Positive intensity was determined by cell staining score: tan, 3 points; brown and yellow, 2 points; light yellow, 1 point; no coloring, 0 points. The sum of the positive percentage and positive intensity was the IHC staining score. According to the above scoring rules, TOP2A expression in HCC was divided into two groups: the low expression group (0–2 points) and the high expression group (3–6 points).
Clinical follow-up
All the 84 paraffin-related patients were followed up for the overall postoperative survival time, which was more than 5 years or until death.
Statistical analysis
All data were analyzed using SPSS 20.0. The expression of TOP2A in HCC and para-cancer tissues and the relationship between TOP2A expression and clinicopathological factors were analyzed by chi-square test. Factors with predictive significance in the univariate model were evaluated in the multivariate Cox regression model. For all trials, the significance level of statistical analysis was P<0.05.
Results
Expression of TOP2A in tumor and para-cancer tissues
Real-time PCR and Western blotting analysis
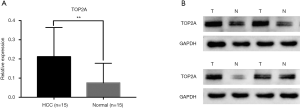
Fifteen pairs of RNA from tumor and para-cancer tissues were extracted and detected by real-time PCR to investigate the difference of TOP2A expression in tumor and para-cancer tissues. mRNA expression of total TOP2A in tumor tissues was significantly higher than that in para-cancer tissues compared with β-actin (P=0.0064) (Figure 1A). Western blotting analysis indicated that TOP2A protein was higher in HCC tumor tissues than in adjacent tissues (Figure 1B).
Immunohistochemical analysis
To explore the correlation between TOP2A expression and clinicopathology at the protein level, paraffin sections of 84 HCC patients from the Nantong Tumor Hospital were collected. Through immunohistochemical analysis, the overexpression of TOP2A in the liver and the distribution of cells in cancer or surrounding tissues were determined (Figure 2). The brown-yellow granules were distributed in the nucleus and a few cytoplasms. The positive rate of TOP2A in HCC was 84.52% (71/84), while the positive rate of TOP2A in para-cancer tissues was 8.33% (7/84), and the expression of TOP2A in HCC was significantly higher than that in para-cancer tissues (chi-square =98.02, P<0.0001) (Table 2).
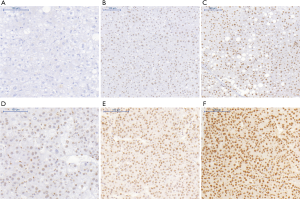
Table 2
| Group | n | TOP2A | χ2 | P value | TOP2A expression strength | χ2 | P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | 0 | 1–2 | 3–4 | 5–6 | ||||||
| HCC | 84 | 13 | 71 | 98.02 | <0.0001 | 13 | 20 | 18 | 33 | 100.6 | <0.0001 |
| Para-cancer | 84 | 77 | 7 | 77 | 2 | 5 | 0 | ||||
TOP2A expression and pathological stage
After joint evaluation by 2 pathologists, 11 patients with grade I, 42 patients with grade II, 29 patients with grade III, and 2 patients with grade IV were classified. There were 53 cases of grade I–II, 27 of which had high TOP2A expression (50.94%). There were 31 cases with grade III−IV, among which 24 cases had high TOP2A expression (77.42%), and the difference between the two groups was significant (P=0.0165) (Table 1).
Relationship between TOP2A expression and tumor size
Among the 84 cases, 50 cases were <5 cm, among which 25 cases had high TOP2A expression (50%). Twenty-four cases were ≥5 and <10 cm, among which 20 cases had high TOP2A expression (83.33%). TOP2A was highly expressed in 6 of the 10 patients (60%) with tumor ≥10 cm. Due to the small number of patients with tumor size greater than or equal to 10 cm, tumor sizes ≥5 cm were counted. A total of 34 patients with tumor size ≥5 cm had high TOP2A expression, among which 26 had high TOP2A expression (76.47%). The results showed that the TOP2A expression of tumor size <5 cm was lower than that of tumor size ≥5 cm, with a significant difference (P=0.0148) (Table 1).
Relationship between TOP2A expression and liver cirrhosis
Among the 84 cases, 21 cases had no obvious cirrhosis, among which 6 cases had high TOP2A expression (28.57%). Sixty-three patients had significant cirrhosis of the liver, of which 45 had high TOP2A expression (71.43%). The results showed that TOP2A expression was higher in patients with cirrhosis than in patients without cirrhosis (P=0.0005) (Table 1).
TOP2A expression and HBV infection
Among all patients, 7 patients had no HBV infection, and 4 of them had high TOP2A expression (57.14%). The remaining 77 cases were all accompanied by hepatitis b virus (HBV) infection, among which 47 cases had high TOP2A expression (61.04%), and there was no significant difference in TOP2A expression between HBV-infected tumors (P=0.8399) (Table 1).
TOP2A expression and overall survival time
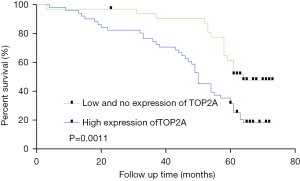
Among all the patients, 37 survived for more than 60 m, and 14 of them had high TOP2A expression (37.84%). The survival time of 47 cases was less than 60 m, among which 37 cases had high TOP2A expression (78.72%), and the difference between the two groups was significant in terms of high TOP2A expression (P<0.0001). Among all the cases, 51 cases had high TOP2A expression, while 33 cases had low or no TOP2A expression (Table 1). Kaplan-Meier survival analysis was performed on the survival time of the two groups. The results showed that the survival time of the group with low TOP2A expression or no TOP2A expression was better than that of the group with high TOP2A expression, and the difference was statistically significant (P=0.0011) (Figure 3).
Clinicopathological features and TOP2A correlation with 5-year survival: COX regression univariate analysis and multivariate analysis
Univariate analysis was performed on the clinical case characteristics and 5-year survival follow-up data of 84 patients, and it was found that cirrhosis and TOP2A expression were respective prognostic factors, while gender, age, tumor grade, HBV infection, and tumor size had no effect on 5-year survival time in this study. After the expression of cirrhosis and TOP2A was substituted into the COX regression multivariate analysis model, it was found that cirrhosis and TOP2A were independent factors affecting the prognosis (Table 3).
Table 3
| Variable | Univariable analysis | Multivariable analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | P | 95% CI | HR | P | 95% CI | ||
| Gender | |||||||
| Male vs. female | 0.976 | 0.945 | 0.493–1.933 | ||||
| Age | |||||||
| ≤60 vs. >60 | 1.182 | 0.571 | 0.663–2.107 | ||||
| Differentiation | |||||||
| Low vs. high | 0.956 | 0.87 | 0.558–1.639 | ||||
| Liver cirrhosis | |||||||
| Negative vs. positive | 4.158 | <0.001 | 1.875–9.221 | 2.778 | 0.016 | 1.209–6.383 | |
| HBV infection | |||||||
| Absent vs. present | 1.029 | 0.951 | 0.411–2.578 | ||||
| Tumor size | |||||||
| <5 vs. ≥5 cm | 1.114 | 0.69 | 0.655–1.893 | ||||
| TOP2A expression | |||||||
| Low vs. high | 3.8 | <0.001 | 2.062–7.003 | 2.819 | 0.001 | 1.490–5.332 | |
Discussion
HCC is the second most deadly malignant tumor in the world. In the Asia-Pacific region, the etiology is mainly related to viral hepatitis, while in China, it is mainly related to HBV infection (13,14). Furthermore, the occurrence and development of HCC is also the common product of a series of specific molecular and/or genetic events, including the abnormal expression of oncogenes and tumor suppressor genes (15-17). HCC has a poor prognosis, but there is still a lack of effective prognostic indicators for use in clinical practice. As a common predictor of chemotherapy efficacy, TOP2A amplification or deletion is significantly correlated with the reactivity of anthracyclines (18,19). It was found that patients with high expression of TOP2A were more sensitive to etoposide and anthracyclines, while those with low expression showed resistance to both drugs (20). In breast cancer, TOP2A expression is closely related to the occurrence and development of tumors and its efficacy on chemotherapy drugs (21,22). It has been noted that the higher the histological grade of breast-invasive ductal carcinoma, the worse the differentiation of tumor cells, and the higher the expression of the TOP2A gene (23). Phelps et al. found that high expression of TOP2A was associated with a poor prognosis of nephroblastoma (24). TOP2A gene expression in esophageal cancer tissues was higher than para-cancer tissues and was closely related to the degree of infiltration, lymph node metastasis, differentiation, and the clinical prognosis of cancer tissues (25). However, the expression of TOP2A in HCC has not been reported. Therefore, TOP2A was selected for further study. By detecting the mRNA level of HCC and paired samples, it was confirmed in the clinical sample collected that TOP2A was significantly highly expressed in the tested tumor tissues (P<0.0001) and negatively correlated with tumor patient prognosis (P=0.002), suggesting that TOP2A has a similar role to proto-oncogene. As it is known, there are three main structural variations of DNA: supercoiling, knotting, and catenation. These topological configurations enable DNA to maintain its compactness without DNA replication and transcription but will hinder the reaction process during transcription or replication. Topoisomerase is responsible for catalyzing and guiding some chemical reactions to release the knot state of DNA. It catalyzes the instantaneous breaking and rejoining of two chains of double-stranded DNA, allowing the chains to pass through each other, thus changing the topological structure of DNA. DNA topoisomerase is the general name of enzymes that catalyze the transformation of DNA topological isomers. Its function is to catalyze the breaking and binding of DNA chains and influence the structure and topology of DNA (26). The topoisomerase Cutting off of the single chain and changing the topological structure is called type I topoisomerase (TOP1), while cutting off of the double chain one is called type II topoisomerase (TOP2). Human TOP2 is composed of 170 kD (TOP2A) and 180 kD (TOP2B). Although they have a high degree of homology in structure, they have significantly different distributions in tissues and cells and are products of different transport processes. TOP2A (topoisomerase II alpha) is one of type II topoisomerase. The encoding gene TOP2A, located on chromosome 17, is high in rapidly proliferating cells, but not expressed in cells that differentiate at the end, and the expression is limited to the cell cycle from the S phase to the G2/M phase. TOP2A protein levels are regulated by multiple systemic factors such as transcription, translation, and post-transcriptional stability of mRNA. TOP2A is mostly located in the nucleus of proliferating cells, and its level in rapid proliferating cells is several times that in resting cells, and it is mainly distributed in the nucleus of the cell. In the cell cycle, the content of TOP2A in the G0/G1 phase is low, the S phase increases, and the G2/M phase reaches a peak, which is the key to regulate the dynamic changes of the nucleic acid spatial structure and control the physiological functions of the nucleic acid. TOP2A is a key enzyme for DNA repair, cell cycle regulation, and chromosome division. TOP2A is also a potential target for many anticancer drugs such as anthracyclines, and its expression level is also related to the efficacy of anthracyclines.
Anthracyclines are used to treat a variety of cancers. However, due to the influence of different molecular mechanisms and signaling pathways of tumors, the mechanisms for this action are still unclear. TOP2A inhibition through inducing cell apoptosis is the focus of the research related to the mechanism of action in anthracyclines. This inhibition causes the release of the DNA superhelix by catalyzing the breaking and rebinding of the double strand of DNA. The enzyme and the double strand of the DNA binding site are then embedded by anthracyclines, which then block DNA reconnection and cannot be repaired, thus killing tumor cells. Therefore, the main mechanism of anthracyclines is inhibiting TOP2A. The overexpression of TOP2A can reflect the proliferation activity of cells and therefore, indicates poor invasion behavior and prognosis (27-29).
In tumors, the expression of TOP2A is proliferation-dependent, and its expression level reflects the proliferation level of the tumor to some extent. Compared with adjacent normal tissues, its contents in esophageal cancer, liver cancer, gastric cancer, and colorectal cancer tend to be highly expressed, and this difference is closely related to the number of S-phase cells (30-32). This suggests that TOP2A may be a specific marker of malignant tumor cells and plays an important biological role in the occurrence and development of malignant tumors. In addition, the high expression of TOP2A in invasive breast cancer is also related to the negative expression of hormone receptor ER and the high expression of ki-67, which affects the disease-free survival and overall survival of breast cancer patients and is regarded as an important prognostic molecular indicator in the development process of breast cancer (33). At the same time, studies have shown that the high expression of TOP2A in tumors is related to malignant biological behaviors such as axillary lymph node metastasis and distant metastasis (34). Combined with the above findings, our results suggest that the high expression of TOP2A in HCC may be closely related to its proliferation and metastasis, and may be used as a biological indicator to predict the prognosis of tumor clinically. Villman et al. (35) also found in their study that the higher the expression of TOP2A the more obvious the malignant biological behavior of tumors and the worse the prognosis of patients. This indicates that TOP2A could be used as an independent prognostic factor in HCC, and the high expression of TOP2A in tumor tissue may predict poor prognosis of patients.
In this study, TOP2AmRNA expression and TOP2A protein expression were detected by RTPCR and IHC in 15 pairs of fresh tumor and paraffin tissues and 84 pairs of paraffin samples, respectively. TOP2A expression in tumor tissues was found to be higher than that in para-cancer tissues, suggesting that TOP2A may be one of the biomarkers for predicting the prognosis of HCC. The study found that the larger the tumor, the higher the degree of cirrhosis, and the higher the pathological grade, the higher the proportion of TOP2A high expression. In the cases with an overall survival time of less than 60 m, the proportion of TOP2A high expression was higher than that of the cases with the overall survival time of more than 60 m. These suggest that TOP2A expression may be related to the clinical progression of HCC.
TOP2A, a biomarker for predicting the prognosis of malignant tumors, has been confirmed in many tumors, including breast cancer, endometrial cancer, and adrenal cancer. So far, it has not been reported as a prognostic marker of HCC. Currently, the clinical factors that influence the prognosis of HCC include high replication of hepatitis virus, cirrhosis, portal vein carcinoma thrombus, tumor size, and number, etc. (36). However, as a single tumor without portal vein tumor thrombus, the prognosis after radical resection varies greatly. There are no biological markers that predict prognosis in such cases. The cases selected in this study were all single lesion without portal vein cancer thrombi. Kaplan-Meier survival analysis found that the overall survival time of patients with high TOP2A expression was shorter than that of patients with low expression. Univariate and multivariate analysis found that both TOP2A and liver cirrhosis were independent predictors of poor prognosis after single HCC radical resection.
Since HCC in China is mostly related to HBsAg, only 7 out of 84 cases in this study did not have HBV infection, giving no indication that HBV infection is related to prognosis. The purpose of this study was to explore the prognostic indicators after radical resection of a single tumor, with limited cases included. Whether our results can be applied to all postoperative HCC cases still requires further prospective study.
Conclusions
The high expression of TOP2A is associated with the invasiveness and poor prognosis of HCC tumors, and, together with liver cirrhosis, can be a prognostic indicator of HCC radical resection.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.46). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the medical ethics committee of Nantong Tumor Hospital (approval number: 2018-024). Informed consent was waived due to the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Song P, Cai Y, Tang H, et al. The clinical management of hepatocellular carcinoma worldwide: A concise review and comparison of current guidelines from 2001 to 2017. Biosci Trends 2017;11:389-98. [Crossref] [PubMed]
- Sempokuya T, Wong LL. Ten-year survival and recurrence of hepatocellular cancer. Hepatoma Res 2019;5:38. [PubMed]
- Selçuk H. Prognostic Factors and Staging Systems in Hepatocellular Carcinoma. Exp Clin Transplant 2017;15:45-9. [PubMed]
- Toyoda H, Kumada T, Tada T, et al. Differences in the impact of prognostic factors for hepatocellular carcinoma over time. Cancer Sci 2017;108:2438-44. [Crossref] [PubMed]
- Moris D, Chakedis J, Sun SH, et al. Management, outcomes, and prognostic factors of ruptured hepatocellular carcinoma: A systematic review. J Surg Oncol 2018;117:341-53. [Crossref] [PubMed]
- Lee SK, Wang W. Roles of Topoisomerases in Heterochromatin, Aging, and Diseases. Genes (Basel) 2019; [Crossref] [PubMed]
- Chen T, Sun Y, Ji P, et al. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene 2015;34:4019-31. [Crossref] [PubMed]
- Lan J, Huang HY, Lee SW, et al. TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol 2014;35:179-87. [Crossref] [PubMed]
- Zhu L, Ma N, Wang B, et al. Clinical analysis of 21-gene recurrence score test in hormone receptor-positive early-stage breast cancer. Oncol Lett 2019;17:5469-80. [PubMed]
- Liu L, Lin J, He H. Identification of Potential Crucial Genes Associated With the Pathogenesis and Prognosis of Endometrial Cancer. Front Genet 2019;10:373. [Crossref] [PubMed]
- Xiao H, Xu D, Chen P, et al. Identification of Five Genes as a Potential Biomarker for Predicting Progress and Prognosis in Adrenocortical Carcinoma. J Cancer 2018;9:4484-95. [Crossref] [PubMed]
- Yu MC, Liu JX, Ma XL, et al. Differential network analysis depicts regulatory mechanisms for hepatocellular carcinoma from diverse backgrounds. Future Oncol 2019;15:3917-34. [Crossref] [PubMed]
- Shirvani-Dastgerdi E, Schwartz RE, Ploss A. Hepatocarcinogenesis associated with hepatitis B, delta and C viruses. Curr Opin Virol 2016;20:1-10. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Zhu Q, Gong L, Liu X, et al. Loss of heterozygosity at D8S262: an early genetic event of hepatocarcinogenesis. Diagn Pathol 2015;10:70. [Crossref] [PubMed]
- Khemlina G, Ikeda S, Kurzrock R. The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer 2017;16:149. [Crossref] [PubMed]
- Gibson EG, King MM, Mercer SL, et al. Two-Mechanism Model for the Interaction of Etoposide Quinone with Topoisomerase IIalpha. Chem Res Toxicol 2016;29:1541-8. [Crossref] [PubMed]
- Mordente A, Meucci E, Martorana GE, et al. Topoisomerases and Anthracyclines: Recent Advances and Perspectives in Anticancer Therapy and Prevention of Cardiotoxicity. Curr Med Chem 2017;24:1607-26. [Crossref] [PubMed]
- Marinello J, Delcuratolo M, Capranico G. Anthracyclines as Topoisomerase II Poisons: From Early Studies to New Perspectives. Int J Mol Sci 2018; [Crossref] [PubMed]
- Zheng H, Li X, Chen C, et al. Quantum dot-based immunofluorescent imaging and quantitative detection of TOP2A and prognostic value in triple-negative breast cancer. Int J Nanomedicine 2016;11:5519-29. [Crossref] [PubMed]
- Nuncia-Cantarero M, Martinez-Canales S, Andres-Pretel F, et al. Functional transcriptomic annotation and protein-protein interaction network analysis identify NEK2, BIRC5, and TOP2A as potential targets in obese patients with luminal A breast cancer. Breast Cancer Res Treat 2018;168:613-23. [Crossref] [PubMed]
- Mitrović O, Cokic V, Dikic D, et al. Correlation between ER, PR, HER-2, Bcl-2, p53, proliferative and apoptotic indexes with HER-2 gene amplification and TOP2A gene amplification and deletion in four molecular subtypes of breast cancer. Target Oncol 2014;9:367-79. [Crossref] [PubMed]
- Phelps HM, Al-Jadiry MF, Corbitt NM, et al. Molecular and epidemiologic characterization of Wilms tumor from Baghdad, Iraq. World J Pediatr 2018;14:585-93. [Crossref] [PubMed]
- Yu Y, Ding S, Liang Y, et al. Expression of ERCC1, TYMS, TUBB3, RRM1 and TOP2A in patients with esophageal squamous cell carcinoma: A hierarchical clustering analysis. Exp Ther Med 2014;7:1578-82. [Crossref] [PubMed]
- Anand J, Sun Y, Zhao Y, et al. Detection of Topoisomerase Covalent Complexes in Eukaryotic Cells. Methods Mol Biol 2018;1703:283-99. [Crossref] [PubMed]
- Baiocchi G, Poliseli FL, De Brot L, et al. TOP2A copy number and TOP2A expression in uterine benign smooth muscle tumours and leiomyosarcoma. J Clin Pathol 2016;69:884-9. [Crossref] [PubMed]
- Labbé DP, Sweeney CJ, Brown M, et al. TOP2A and EZH2 Provide Early Detection of an Aggressive Prostate Cancer Subgroup. Clin Cancer Res 2017;23:7072-83. [Crossref] [PubMed]
- Wang TL, Ren YW, Wang HT, et al. Association of Topoisomerase II (TOP2A) and Dual-Specificity Phosphatase 6 (DUSP6) Single Nucleotide Polymorphisms with Radiation Treatment Response and Prognosis of Lung Cancer in Han Chinese. Med Sci Monit 2017;23:984-93. [Crossref] [PubMed]
- Sudan S, Rupasinghe HP. Quercetin-3-O-glucoside induces human DNA topoisomerase II inhibition, cell cycle arrest and apoptosis in hepatocellular carcinoma cells. Anticancer Res 2014;34:1691-9. [PubMed]
- Wu Z, Zhao Y, Zhang Y, et al. HY-2, a novel DNA topoisomerase II inhibitor, induces G2/M cell cycle arrest in HCT-116 cells. J Chemother 2014;26:342-7. [Crossref] [PubMed]
- Jain CK, Roychoudhury S, Majumder HK. Selective killing of G2 decatenation checkpoint defective colon cancer cells by catalytic topoisomerase II inhibitor. Biochim Biophys Acta 2015;1853:1195-204. [Crossref] [PubMed]
- Nakagawa M, Bando Y, Nagao T, et al. Expression of p53, Ki-67, E-cadherin, N-cadherin and TOP2A in triple-negative breast cancer. Anticancer Res 2011;31:2389-93. [PubMed]
- Fountzilas G, Valavanis C, Kotoula V, et al. HER2 and TOP2A in high-risk early breast cancer patients treated with adjuvant epirubicin-based dose-dense sequential chemotherapy. J Transl Med 2012;10:10. [Crossref] [PubMed]
- Villman K, Sjostrom J, Heikkila R, et al. TOP2A and HER2 gene amplification as predictors of response to anthracycline treatment in breast cancer. Acta Oncol 2006;45:590-6. [Crossref] [PubMed]
- Fujiwara N, Friedman SL, Goossens N, et al. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 2018;68:526-49. [Crossref] [PubMed]


