
Applications of hyperspectral imaging in the detection and diagnosis of solid tumors
Introduction
Solid tumor
Cancer, also referred to as a malignant tumor, has become the leading cause of death in China since 2010 (1). Clinically, tumors can be classified into solid tumors and non-solid tumors. Non-solid tumors, such as leukemias, cannot be effectively detected by medical imaging or palpation. However, for solid tumors (2,3), imaging tests are a critical tool for diagnosis.
For many types of cancers, noticeable symptoms do not show until middle or late stage, which makes it difficult to early diagnosis and treatment (4). The diagnosis of cancer is to locate the primary tumors and determine their histopathological types and the stages of invasion and metastasis. Until now, biopsy histopathological analysis remains the gold standard for cancer diagnosis, which is time- and cost-consuming (5). Medical imaging methods are commonly performed throughout the diagnostic procedure to help detect and localize abnormal structures.
Routine medical imaging methods
Medical imaging can be divided into two main categories (6,7): (I) biomedical micrography, which mainly targets the microstructures of organisms, and (II) anatomical macroscopy, also referred to as modern medical imaging, which focuses on the anatomical structure and function of the human body. At present, the most widely used medical imaging techniques are X-ray, computed tomography (CT), magnetic resonance imaging (MRI), ultrasonography, radionuclide imaging, and optical imaging (8,9).
From the perspective of imaging depth and spatial resolution, some of the aforesaid imaging methods are listed and compared (Figure 1). Both CT and MRI are capable of imaging the depth of full body. But their imaging resolution is limited to approximately 1 mm (8,10,11). Microscopy imaging, as an optical imaging technique, shows the highest resolution at 1 µm. However, its detection depth can only reach about 100 µm (3). Various methods have been developed to overcome these deficiencies. For example, an optical imaging method called optical coherence tomography (OCT) could reach a deeper place with light sources of different bandwidths (12). Ultrasonic imaging stands in the middle, achieving a 10–100 µm resolution and an imaging depth of a few centimeters (9). By comparison, optical imaging typically has high spatial resolution; however without additional improvement, it could only reach a depth of a few millimeters due to the propagation limit of light when it travels through tissues (9,13). Hyperspectral imaging (HSI) falls within the scope of optical imaging. Figure 1 illustrates the imaging depth and resolution of major medical imaging methods. An extra axis for the spectral range is added to better present the distinction between HSI and other methods.
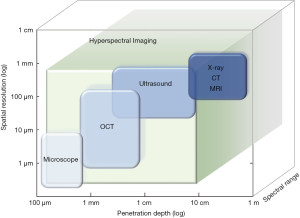
All these medical imaging methods have their own scopes in clinical applications. In most cases, they are complementary and synergistic, rather than substitutive. Combinations of different medical imaging methods can help detect the initial pathological changes in human tissues more accurately and provide means for early diagnosis and treatment than single imaging modalities (8,14,15). A broader spectral band and higher spectral resolution provide richer information for tumor diagnosis and detection. HSI is a promising technique that prossesses these qualities and thus provides a new viewpoint in the medical imaging field.
HSI
Spectral imaging, also known as imaging spectroscopy, is a new technology that combines imaging and spectrum to obtain spatial and spectral information from a scene simultaneously. HSI is a subcategory of spectral imaging that continuously captures tens to hundreds of spectral bands. It collects information from three dimensions–two spatial (x, y) and one spectral (λ), resulting in a (x, y, λ) dataset, which is typically referred to as a data cube (16) (Figure 2). In brief, HSI captures a set of spectrally resolved images, or spatially resolved spectra. Although HSI originated from remote sensing (17,18), widespread applications have been found in biomedical imaging (5,19), food quality assessment (20,21), surveillance (22), etc. Due to the diverse combinations of imaging principles (e.g., spatial scanning, spectral scanning, and snapshot) and light splitting methods (e.g., dispersion, filtering, and modulation), there are many ways to implement a HSI system. Readers with interest can refer to relevant studies (16,18,23-25).
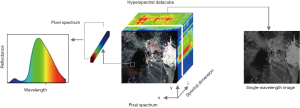
HSI is a non-invasive and rapid technique that can be applied to solid tumors. HSI is able to capture images under different spectral bands (e.g., visible, infrared, ultraviolet band) at the same time in a non-invasive manner (17), while common optical imaging methods could only obtain a limited number of spectral bands. The spatial information of HSI images shows the size and shape of target objects, while the spectral information reveals composition-specific information under hundreds of wavelengths (16). The specific spectral information is considered as spectral fingerprints that reflects the chemical composition and content of different substances. When disease develops, the optical and pathological properties of tissues will change accordingly (5,19). Spectral features induced by these changes can be captured by HSI systems and then transformed into quantitative diagnostic information. Because of the biological tissue structure and chromophores (26,27), the light will go through a series of scattering and absorption when travelling through biological tissues. Scattering occurs more often than absorption, becoming the dominant phenomenon when light interacts with tissues. The absorption spectrum reflects the degree of angiogenesis and metabolism (26). The reflected spectrum reveals the scattering and absorption information of tissue components. HSI detects cancers by measuring the absorption, fluorescence emission or reflectance spectrum of tissues. Additionally, there exist some multi-model designs, such as a combined reflection and fluorescence imaging system (28).
Although the resolution and spectral properties of HSI are outstanding, we still need to mention the drawbacks of HSI methods, namely, the imaging depth. Light entering tissues is affected by absorption and scattering; therefore, it has a propagation limit, i.e., a limit of optical penetration depth. The optical penetration depth varies with tissue composition and wavelength (29). Although near-infrared (NIR) light performs better than that in the visible band, it still only reaches millimeter-level depths in skin tissue (30). This limitation explains why optical imaging methods are mostly used on slices or shallow surfaces (31). Current in vivo tumor detection by HSI is mainly focused on exterior organs, such as the head, neck, skin, and other organs that can be reached by endoscopy, colposcopy, gastroscopy, or laparoscopy (32-34).
HSI could help us extend our vision to hundreds of invisible bands. Recently, HSI has become increasingly popular in medical fields for its label-free and real-time imaging features. This review will investigate the use of HSI in different circumstances, distinguishing tumor from healthy tissue in vitro and in vivo.
HSI applications in tumor detection
The applications of HSI in tumor detection can be categorized into three groups (5,19,35): in vitro imaging for pathological samples, in vivo imaging on shallow surfaces, and intraoperative applications. Currently, histopathological examination is still regarded as the gold standard for diagnosis. It is time-consuming and strongly dependent upon experience. We will investigate the use of HSI as a new tool to assist pathologists in label-free quantitative estimation (36) and to differentiate tumor tissues in a surgical setting.
In vitro tumor detection of tissue samples
HSI detection of tissue samples has been used to study histiocyte morphology and the behavior of tumors. The tissue samples can be fresh or processed, such as frozen, formalin-fixed paraffin-embedded or stained. They also have many forms, like tissue biopsy sections (37-39), tissue microarray (TMA) (40), and cells (41,42).
Most of the histological characteristics are closely related to tumor development, so normal, benign and malignant tumors can be classified accordingly (43-45). This involves a direct morphological observation on processed biopsy slices, or on an ultrastructure (e.g., nucleus, stroma) under a microscope. The general method is to conduct microscopic inspection of treated samples and define the stage of diseases according to changes in tissue/cell structures and distributions. Taking advantage of the composition-specific spectral markers, HSI can directly distinguish different cell types and tissue patterns in a label-free way (46). In addition, hyperspectral images contain much more information than common red, green, and blue (RGB) three-color images, leading to an improvement in the accuracy of diagnosis.
In the microenvironment, molecular changes in some proteins, nucleic acids and glycogens also indicate different stages of tumors in non-invasive diagnosis and targeted therapy (47). Other important markers in the microenvironment are tissue blood volume and blood oxygen level. During the development of tumors, the consumption of oxygen is far greater than the supply, causing hypoxia in the tumors (48). HSI also captures these subtle changes in the microenvironment.
In advance, we can see that much of the research on histopathological HSI diagnosis is based on chemical imaging in the visible and NIR wave bands (30,49). Chemical imaging is a special case of HSI, covering only the disciplines of chemical interest (50). Specifically, infrared chemical imaging combined with machine learning algorithms are widely used to identify normal and abnormal tissues with satisfying sensitivity and specificity. The infrared spectral features show great importance in revolutionizing the diagnosis of histopathology (50,51). Fourier transform infrared spectroscopy (FTIR), the most commonly used method, provides a label-free method for histopathological examination of cancers (52,53).
Here we briefly summarize some HSI-related research carried out on tumors of the liver, breast, and the gastrointestinal system. Although this review cannot cover all types of tumors, HSI has exhibited great potential in the diagnosis of other solid tumors.
Liver tumors
The most prevalent malignant primary liver tumor is hepatocellular carcinoma (HCC) that usually develops from the progression of cirrhosis. In terms of the pathological observation, many HSI-based diagnosis studies are performed on tissue specimens and cells.
Staining is time-consuming. With the help of HSI, it is possible to observe specimens without staining or fixation. A study by Hashimoto et al. (54) conducted a computer-aided diagnosis on liver pathological samples which were hematoxylin and eosin (H&E) stained. As the cytoplasm in the RGB image is almost identical to the fiber, the classification result shows low accuracy. Applying HSI to the same samples, the accuracy improvement has reached to 24% for fibers and 5% for cytoplasm. In the stroma microenvironment, fibrosis can also play a role as an auxiliary diagnostic spectral marker (55).
FTIR measurement is useful in distinguishing cancerous tissues. It can even indicate the initial cancer-like changes within an area classified as normal tissue. A study by Coe et al. (56) used infrared imaging spectroscopy of human liver tissue to identify and characterize a liver metastasis originating from the breast without fixation or dehydration. The results show differences between tumors and non-tumors with the help of lipid and protein changes. Tissue areas containing fewer lipids are more likely to be tumor areas. The secondary structure of proteins has also been proven to undergo substantial changes when transitioning from non-tumor to tumor areas. Kochan et al. (25) compared two of the most common FTIR measurement modes with normal and cancerous canine liver histiocytic sarcoma tissue in NIR bands. Principal components analysis (PCA), unsupervised hierarchical clustering analysis (HCA) and k-means clustering methods were adopted. The results show that the main difference between normal tissue and cancer tissue lies in the changes in deoxyribonucleic acid (DNA) and carbohydrate content. The study demonstrated the tumor areas have a much higher content of DNA (through increased intensity of the band at 964 cm−1) along with substantially reduced amounts of carbohydrates. This could act as a potential marker for tumor delineation. Combining FTIR with microscopy, the detection can be extended to the cell level. HCA can also be used here to distinguish cancerous and normal cells in several cancer cell lines as well as normal human liver cells (57).
Breast tumors
Breast cancer is one of the most common malignant tumors in females (1). Prognosis and survival rates vary greatly depending on the tumor type, size, grade, stage, and lymph node involvement (58). To date, surgeons must wait for the results of histopathological diagnosis to make a decision for or against mastectomy (5).
Differences in the reflectance and transmittance spectra of tissue components under visible and NIR bands can be used as spectral fingerprints for different tissues, including normal, benign, and malignant tissue samples (5). Morphological changes, such as the nucleus of breast cancer cells, are classified into various tumor stages (45). Measuring both spectral difference and morphological changes, studies achieved better classification accuracy with the aid of HSI (37,40-42).
Recent developments in broadly tunable lasers in the mid-infrared range is considered as the most promising potential candidate for next generation microscopes (59). A mid-to-far-infrared portion of the electromagnetic spectrum can be obtained using quantum cascade lasers (QCLs) as a light source. This approach not only improves throughput but also increases the acquisition speed, which is crucial for clinical translation. Pilling et al. (40) introduced a case of FTIR microscopy and QCL for wide-field infrared imaging. They used a breast cancer TMA comprising 207 different patient samples and implemented a QCL to perform continuous spectral imaging in the band from 912 to 1,800 cm-1, which accurately distinguished malignant and nonmalignant stroma spectra with high sensitivity (93.56%) and specificity (85.64%). Additionally, a study by Fabian et al. (37) developed an automated classifier built by artificial neural networks to separate four classes of breast tissue based on their spectra: fibroadenoma, ductal carcinoma in situ, connective tissue and adipose tissue. Same for diagnosis, Bird et al. (41) detected metastatic breast cancer cells. They reproduced the histological architecture of axillary lymph nodes accurately. As a result, superior sensitivity for cellular abnormality and tissue classification were achieved. Verdonck et al. (42) proved that infrared imaging can serve as a diagnostic tool for identifying morphological and molecular pathologic alterations characteristics in breast epithelial cells and the stroma. A good combination of infrared imaging and an unsupervised/supervised model efficiently identified the main cell types present in breast tissue sections, i.e., epithelial cells, lymphocytes, connective tissue, vascular tissue and erythrocytes.
Research generally focuses on the features of parenchymal and epithelial cells, given that their dysplasia and progression are often the main reason for the development of carcinoma. But, recently, researchers have begun to turn their attention to the cell micro-environment of tumors, the adjacent regions of stroma and fibrosis. Stromata in the vicinity of the tumor exhibit distinct spectral characteristics (42). Researchers hope to detect cancer progression from the microenvironment to improve cancer diagnosis in pre-invasive stages (36,40,60). Much collagenous stromata can be found in the lesions of breast tissues. Disorientation of the collagen fiber network is a common phenomenon during cancer invasion process. It suggested spectral features present in the 1,700–1,600 cm-1 range could act as spectral markers to identify the collagen degradation, which could possibly be used to monitor the invasive processes (61).
Gastrointestinal tumors
Given the low diagnostic rate of traditional esophagogastroduodenoscopy (EGD) examinations and endoscopic systems, Kiyotoki et al. (62) adopted HSI as a new method for gastric cancer detection. They performed HSI on 16 tumor samples from endoscopic resections or surgeries and tested on 726 wavelengths through the established training algorithm and an enhanced image processing. Differences in spectral reflectance between normal and cancerous mucosa were identified. Based on the results of that study, Goto et al. (63) studied the spectral reflectance of gastric tumor tissue and normal mucosa tissue in 81 bands between 400 and 800 nm. The study showed that the 770 nm wavelength and 1/4 cut-off value of spectral reflectance are optimal in detecting gastric tumors with higher accuracy. Additionally, in vitro, Akbari et al. (64) performed HSI infrared analysis on gastric tumors in the 1,000–2,500 nm band. The results indicate that the 1,226–1,251 and 1,288–1,370 nm bands can serve as criteria for distinguishing gastric tumor tissues and non-gastric tumor tissues. Tumor sizes smaller than 0.5 mm can also be detected. These results are still maintained even when the depth of the tumor is less than 2–3 mm with mucosal and normal tissue coverage. Compared to the laser-excited fluorescence used by many research groups to detect cancer, there is no need to inject fluorescent materials for HSI. Since the detection is based on individual pixels, the size and shape of tumor tissues will not affect the classification result. A series of studies (34,62) correct the spectral reflectance and improves the accuracy of the detection. The disadvantage of this procedure is that the samples are all measured in vitro. Therefore, the expectation is that this procedure will be extended to in vivo studies.
In summary, HSI examinations are mostly performed by microscopic infrared imaging. The aforementioned three cases of specific tumor diagnoses have mostly covered the application scope. Histopathological diagnosis can be carried out in vivo or in vitro. There are ways to conduct in vivo detection through endoscopes (33,34) in the form of multi-spectrum detection, i.e., using a few discrete spectra. In vitro, the most welcomed imaging method is FTIR microscopic imaging. Biomedical samples treated in TMA format with a wide-field light source, e.g., QCLs, are used to obtain a higher throughput. Optical diagnosis can be carried out under a traditional morphological analysis with the aid of infrared imaging, or under an advanced way of label-free detection based on the composition-related spectral signatures. Recently, the focus of research has shifted slightly to elements of the microenvironment, such as the stroma. Only three specific cases are presented here, but we can see the similarity in all solid tumors. Previous studies also used HSI as a diagnostic tool in cancers of the prostate (34,65-67), lung (43,68-70), cervix (71), colon (39,44,67,72-76), kidney (77), brain (78,79), skin (80,81), oral cavity (82), and bladder (83), etc.
Non-invasive tumor detection on shallow surfaces
Due to the limits on optical penetration depth, HSI is applied to some openly accessible sites such as the skin, eye mask, cervix, tongue, and microvessel. But it can also be used for some interior organs with the help of endoscopy (32,84).
As the largest organ of the body, the skin is the most studied research subject. Skin imaging could differentiate different outgrowths, such as common nevus, spitz nevus, malignant melanoma, basal cell carcinoma and psoriasis (85,86). Previously, dermoscopy was the fastest device to obtain skin images. Recently, the use of HSI for the diagnosis of skin cancer has shown excellent performances (87,88). Figure 3 shows general steps of HSI body surface inspection. The principle is to obtain different reflectance and absorption characteristics of different skin lesion areas under different wavelengths of light.
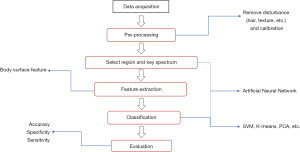
The overall setup of HSI is shrinking, allowing some HSI systems to be easily integrated on portable devices or even smartphones. MelaFind (89) is a non-invasive spectral imaging tool, developed by the MELA Sciences company. It uses light from the visible to NIR wavelengths to evaluate skin lesions up to 2.5 mm beneath the skin, providing additional information to dermatologists during melanoma skin examinations (90). The MelaFind pivotal study showed a 98.3% sensitivity by identifying 172 out of 175 melanomas and high-grade lesions. Furthermore, researchers developed a smartphone-based multispectral imaging system for the early skin diagnosis and quantitative prognosis monitoring at home (91).
Non-invasive HSI detection has also been reported for other tumor types, such as head and neck cancers (92), including tumors from the lips, mouth, tongue, nose, throat, and lymph nodes (5,32,93), etc. These tumors all occur in the epithelial areas. For instance, a series of studies performed feature extraction and classification of the textures on the tongue surface, and then linked those features with oral cancer to study their malignant changes (94).
HSI can also be used for interior organs, such as the stomach (32,95), colon (32) and cervix (33,71), with the help of colposcopy, endoscopy, gastroscopy and so on. Colposcopy is the most commonly used method for conducting cervical intra-epithelial neoplasia tests. HSI helps to categorize and quantify the stages and precancerous stages of cervical cancer (34). Today, combined HSI and endoscopy systems have already reached a video frame rate for a better intraoperative use (96,97).
HSI in surgical applications
For the translation of the technique to an in vivo setting, where it will be used during surgery, better real-time, in vivo, and label-free performances are favorable. To date, traditional imaging methods, such as CT and MRI, help to confirm tumor locations prior to appropriate resection surgeries. However, the specific locations, number and sizes of tumors hiding within organs may be uncertain (98). During the surgery, the experience and ability of surgeons, to perform visual diagnosis, eye-hand coordination and tactile feedback tend to be dominant. Hence, there is a growing need for intraoperative imaging apparatus. Because of the in vivo, real time, and label-free characteristics, HSI has received much attention in peri-operative application and therapy assessment. HSI has shown its promising ability to distinguish normal and cancerous tissues in nearly real time in surgical oncology. HSI images, together with some sophisticated classification algorithms, can monitor the surgical microenvironment, perform tumor delimitation, implement surgery navigation, identify important adjacent structures, detect remaining tumor cells, and finally assist surgeons in making accurate surgical decisions (19,99).
Visualizing the surgical environment
HSI can differentiate other normal tissues such as vessels, connective tissues, and muscles, and assess dynamic organ perfusion without interfering with the ongoing surgery (34,100). In clinical use of HSI, microvasculature and tumor hypoxia show the prognostic value in surgery (101). Research on real-time imaging of tissue oxygenation was carried out (102). A study by Holzer et al. (103) demonstrated that HSI could be used as a non-invasive and highly sensitive tool for real-time monitoring of renal oxygenation and blood flow during open partial nephrectomy for renal cortical tumors in humans. In another work, Tetschke et al. (104) reported that HSI can detect and monitor the oxygen saturation levels and water characteristics of the kidney tissue in preservation process. So, HSI has been proved to be a beneficial tool for protecting the functioning of organs and monitoring surgical environment. To ensure the real-time performance, intraoperative applications have high expectations of imaging speed. Zuzak et al. (102) verified a digital light processing technology based HSI system and achieved a near video speed of three chemically encoded images per second. A study (105) by Pichette et al. showed that HSI could capture the brain hemodynamic response at a video speed with molecular specificity, which allows for a better understanding of neurovascular coupling and cerebral metabolism.
Intraoperative navigation for minimal injury
Large wounds are susceptible to infection and are not conducive to wound healing. Though laparoscopy can help surgeons perform minimally invasive surgery, it is still based on two-dimensional images and a lack of tactile feedback, similar to traditional image-guided methods. New trials are needed for anatomical navigation. HSI has made some progress in this field. It could improve intraoperative anatomic localization and the identification of some critical features during laparoscopy (100,106). Zuzak et al. (107) used a NIR HSI system combined with a conventional laparoscopy to show a real-time, in vivo display of tissues, status, and anatomical structures during a cholecystectomy. The results showed that even the gallbladder could be differentiated from the liver according to the chemical composition of different tissues. It further improved the contrast of different tissues in the original laparoscopic images. The minimal injury with HSI reduces the chance of wound infections and facilitates faster patient recovery.
Tumor margin assessment
HSI acts as an intraoperative aid tool for the assessment of tumor margins during and after resections (5). Studies are first carried out on ex-vivo surgical specimens, after which label-free HSI can be used for rapid and objective assessments of tumor margins (108). Researchers designed surgical protocols to collect and image surgical specimens within several minutes after resection. Then follows the gold-standard examination of staining or immunohistochemical observation by a pathologist which occurs days after the resection (109,110). The purpose of the examinations is to ascertain whether any tumor tissues or cells are near the edge of the excised specimen, indicating that the resection may not have been successful. The feasibility of using HSI to detect and delineate cancers in fresh, surgical specimens from patients has been investigated for head and neck cancer (108,111), breast cancer (29,109,112), and brain cancer (113,114), etc. Studies established a HSI spectral database of cancer and normal tissues from numerous patients and a variety of tissue types from different anatomical sites. Histopathological results served as the labels (108,115). After that, prediction models and algorithms were built and optimized to predict cancerous tissue during the surgical resection (31,92,114,116,117). In addition to the assessment of excised specimens, the importance of direct examination of residual tumors left in the surgical cavity has also been addressed (5,118,119). Therefore, HSI provides a powerful way to ensure tumor margins in a more time and cost-effective manner, thereby reducing the suffering of the patients and the workload of the surgeons.
Conclusions
In summary, HSI combines the power of imaging and spectroscopy, and provides both spectral and spatial information. Spatial information helps us see the morphologies and changes intuitively, while the spectral information acts as fingerprints reflecting component-specific information. Thus far, HSI has helped with the diagnosis, micro-environmental monitoring, and margin assessment of solid tumors in vitro and in vivo. The composition-related spectral features pave a way for non-staining and label-free examinations. The limitation of HSI lies in its optical penetration depth. But with the help of endoscopy, the observations from HSI can be extended from the shallow surfaces to the interior organs. Additionally, by combining HSI with other techniques, advanced hybrid systems may have better performance. With the rich spectral information and remarkable achievement, HSI will show its place in the medical field.
Suggested future research
There are several directions for future research on this topic. We suggest a few that we feel are particularly important here. (I) The limit of penetration depth. There is still a need for studies to improve the imaging depth by combining it with other techniques, e.g., the optoacoustic imaging systems. (II) Studies on hardware improvement are encouraged. HSI systems have different performances in imaging speed, spatial resolution, spectral range and resolution. It is worth mentioning that HSI devices are becoming cheaper and smaller, facilitating the widespread production and application (120). The real-time acquisition of the three-dimensional hyperspectral data becomes a challenge. (III) HSI algorithms sometimes have poor interpretation of the relationship between spectral signatures and tissue components. There is no specific standard to evaluate their results. (IV) The amount of hyperspectral data is often unwieldy, i.e., the three-dimensional data cube is much larger than the data we used to get. Therefore, it is significant to develop algorithms suitable for hyperspectral data rather than using traditional data processing methods. (V) To better develop the HSI research in the medical field, public medical hyperspectral image datasets are needed for evaluating different medical HSI methods. Requirements for the establishment of spectral databases for important molecular markers have also been put forward. (VI) There is a mismatch between theory and current practice. Most HSI systems have not been widespread in clinical use. Procedures for sample preparation, pre-processing and establishing the image capture system parameters are not consistent, thus preventing the large-scale verification required in the medical domain. We hope to provide surgeons with reliable data, convenient equipment and real-time results through HSI technology. Standards and additional verification tests are required to meet the safety requirements before being put into clinical practice.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.53). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Brown JM, Giaccia AJ. The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res 1998;58:1408-16. [PubMed]
- Danese E, Montagnana M, Lippi G. Circulating molecular biomarkers for screening or early diagnosis of colorectal cancer: which is ready for prime time? Ann Transl Med 2019;7:610. [Crossref] [PubMed]
- Chakraborty S, Rahman T. The difficulties in cancer treatment. Ecancermedicalscience 2012;6:ed16. [PubMed]
- Lu G, Fei B. Medical hyperspectral imaging: a review. J Biomed Opt 2014;19:10901. [Crossref] [PubMed]
- Weissleder R, Nahrendorf M. Advancing biomedical imaging. Proc Natl Acad Sci U S A 2015;112:14424-8. [Crossref] [PubMed]
- Wang C, Wang Z, Zhao T, et al. Optical molecular imaging for tumor detection and image-guided surgery. Biomaterials 2018;157:62-75. [Crossref] [PubMed]
- Pian Q, Wang C, Chen X, et al. Multimodal biomedical optical imaging review: towards comprehensive investigation of biological tissues. Curr Mol Imaging 2014;3:72-87. [Crossref]
- Chen Y, Yuan S, Wierwille J, et al. Integrated optical coherence tomography (OCT) and fluorescence laminar optical tomography (FLOT). IEEE J Sel Top Quant 2010;16:755-66. [Crossref]
- Weber WA, Figlin R. Monitoring cancer treatment with PET/CT: does it make a difference? J Nucl Med 2007;48:36S-44S. [PubMed]
- Dayakshini S, Kamath S, Rajagopal KV, et al. Medical imaging techniques and computer aided diagnostic approaches for the detection of breast cancer with an emphasis on thermography - a review. IJMEI 2016;8:275-99. [Crossref]
- Zysk AM, Nguyen FT, Oldenburg AL, et al. Optical coherence tomography: a review of clinical development from bench to bedside. J Biomed Opt 2007;12:051403. [Crossref] [PubMed]
- Fujimoto JG, Pitris C, Boppart SA, et al. Optical coherence tomography: an emerging technology for biomedical imaging and optical biopsy. Neoplasia 2000;2:9-25. [Crossref] [PubMed]
- Stoffels I, Morscher S, Helfrich I, et al. Metastatic status of sentinel lymph nodes in melanoma determined noninvasively with multispectral optoacoustic imaging. Sci Transl Med 2015;7317ra199.
- Taruttis A, Ntziachristos V. Advances in real-time multispectral optoacoustic imaging and its applications. Nat Photonics 2015;9:219-27. [Crossref]
- Gao L, Smith RT. Optical hyperspectral imaging in microscopy and spectroscopy - a review of data acquisition. J Biophotonics 2015;8:441-56. [Crossref] [PubMed]
- Eismann M T. Hyperspectral remote sensing. Bellingham: SPIE, 2012.
- Manolakis, Dimitris G., Ronald B. Lockwood, and Thomas W. Cooley. Hyperspectral imaging remote sensing: physics, sensors, and algorithms. Cambridge University Press, 2016.
- Li Q, He X, Wang Y, et al. Review of spectral imaging technology in biomedical engineering: achievements and challenges. J Biomed Opt 2013;18:100901. [Crossref] [PubMed]
- Liu Y, Pu H, Sun DW. Hyperspectral imaging technique for evaluating food quality and safety during various processes: A review of recent applications. Trends Food Sci Tec 2017;69:25-35. [Crossref]
- Siche R, Vejarano R, Aredo V, et al. Evaluation of food quality and safety with hyperspectral imaging (HSI). Food Eng Rev 2016;8:306-22. [Crossref]
- Fossi AP, Ferrec Y, Roux N, et al. Miniature and cooled hyperspectral camera for outdoor surveillance applications in the mid-infrared. Opt Lett 2016;41:1901-4. [Crossref] [PubMed]
- Wu J, Xiong B, Lin X, et al. Snapshot Hyperspectral Volumetric Microscopy. Sci Rep 2016;6:24624. [Crossref] [PubMed]
- Heist S, Zhang C, Reichwald K, et al. 5D hyperspectral imaging: fast and accurate measurement of surface shape and spectral characteristics using structured light. Opt Express 2018;26:23366-79. [Crossref] [PubMed]
- Kochan K, Heraud P, Kiupel M, et al. Comparison of FTIR transmission and transfection substrates for canine liver cancer detection. Analyst 2015;140:2402-11. [Crossref] [PubMed]
- Welch AJ, van Gemert MJ, Star WM. Definitions and Overview of Tissue Optics. In: Welch AJ, Van Gemert MJ (eds). Optical-Thermal Response of Laser-Irradiated Tissue. Springer, Dordrecht, 2010:27-64.
- Kim A, Wilson BC. Measurement of Ex Vivo and In Vivo Tissue Optical Properties: Methods and Theories. In: Welch A, van Gemert M (eds). Optical-Thermal Response of Laser-Irradiated Tissue. Springer, Dordrecht, 2010:267-319.
- Novikova IV, Smallwood CR, Gong Y, et al. Multimodal hyperspectral optical microscopy. Chem Phys 2017;498:25-32. [Crossref]
- Kho E, de Boer LL, Post AL, et al. Imaging depth variations in hyperspectral imaging: Development of a method to detect tumor up to the required tumor-free margin width. J Biophotonics 2019;12:e201900086. [Crossref] [PubMed]
- Bashkatov AN, Genina EA, Kochubey VI, et al. Optical properties of human skin, subcutaneous and mucous tissues in the wavelength range from 400 to 2000 nm. J Phys D Appl Phys 2005;38:2543. [Crossref]
- Lu G, Halig L, Wang D, et al. Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging. J Biomed Opt 2014;19:106004. [Crossref] [PubMed]
- Regeling B, Thies B, Gerstner AO, et al. Hyperspectral Imaging Using Flexible Endoscopy for Laryngeal Cancer Detection. Sensors (Basel) 2016;16:1288. [Crossref] [PubMed]
- Ferris DG, Lawhead RA, Dickman ED, et al. Multimodal hyperspectral imaging for the noninvasive diagnosis of cervical neoplasia. J Low Genit Tract Dis 2001;5:65-72. [PubMed]
- Ogihara H, Hamamoto Y, Fujita Y, et al. Development of a gastric cancer diagnostic support system with a pattern recognition method using a hyperspectral camera. J Sensors 2016;2016:1803501. [Crossref]
- Halicek M, Fabelo H, Ortega S, et al. In-Vivo and Ex-Vivo Tissue Analysis through Hyperspectral Imaging Techniques: Revealing the Invisible Features of Cancer. Cancers (Basel) 2019;11:756. [Crossref] [PubMed]
- Nazeer SS, Sreedhar H, Varma VK, et al. Infrared spectroscopic imaging: Label-free biochemical analysis of stroma and tissue fibrosis. Int J Biochem Cell Biol 2017;92:14-7. [Crossref] [PubMed]
- Fabian H, Thi NA, Eiden M, et al. Diagnosing benign and malignant lesions in breast tissue sections by using IR-microspectroscopy. Biochim Biophys Acta 2006;1758:874-82. [Crossref] [PubMed]
- Pounder FN, Reddy RK, Bhargava R. Development of a practical spatial-spectral analysis protocol for breast histopathology using Fourier transform infrared spectroscopic imaging. Faraday Discuss 2016;187:43-68. [Crossref] [PubMed]
- Kuepper C, Kallenbach-Thieltges A, Juette H, et al. Quantum Cascade Laser-Based Infrared Microscopy for Label-Free and Automated Cancer Classification in Tissue Sections. Sci Rep 2018;8:7717. [Crossref] [PubMed]
- Pilling MJ, Henderson A, Gardner P. Quantum Cascade Laser Spectral Histopathology: Breast Cancer Diagnostics Using High Throughput Chemical Imaging. Anal Chem 2017;89:7348-55. [Crossref] [PubMed]
- Bird B, Bedrossian K, Laver N, et al. Detection of breast micro-metastases in axillary lymph nodes by infrared micro-spectral imaging. Analyst 2009;134:1067-76. [Crossref] [PubMed]
- Verdonck M, Denayer A, Delvaux B, et al. Characterization of human breast cancer tissues by infrared imaging. Analyst 2016;141:606-19. [Crossref] [PubMed]
- Bird B, Miljković MS, Remiszewski S, et al. Infrared spectral histopathology (SHP): a novel diagnostic tool for the accurate classification of lung cancer. Lab Invest 2012;92:1358-73. [Crossref] [PubMed]
- Masood K, Rajpoot N. Texture based classification of hyperspectral colon biopsy samples using CLBP, 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE, Boston, 2009:1011-4.
- Boucheron LE, Bi Z, Harvey NR, et al. Utility of multispectral imaging for nuclear classification of routine clinical histopathology imagery. BMC Cell Biol 2007;8:S8. [Crossref] [PubMed]
- Berisha S, Lotfollahi M, Jahanipour J, et al. Deep learning for FTIR histology: leveraging spatial and spectral features with convolutional neural networks. Analyst 2019;144:1642-53. [Crossref] [PubMed]
- Riethdorf S. Detection of microRNAs in circulating tumor cells. Transl Cancer Res 2018;7:S197-208. [Crossref]
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Hermes M, Morrish RB, Huot L, et al. Mid-IR hyperspectral imaging for label-free histopathology and cytology. J Optics 2018;20:023002. [Crossref]
- Pilling M, Gardner P. Fundamental developments in infrared spectroscopic imaging for biomedical applications. Chem Soc Rev 2016;45:1935-57. [Crossref] [PubMed]
- Diem M, Papamarkakis K, Schubert J, et al. The infrared spectral signatures of disease: extracting the distinguishing spectral features between normal and diseased states. Appl Spectrosc 2009;63:307A-318A. [Crossref] [PubMed]
- Kochan K, Baranska M, Maslak E. Biomedical Application of Raman and FT-IR Spectroscopies: Label‐Free Imaging of Liver Insult. Encyclopedia of Analytical Chemistry: Applications, Theory and Instrumentation 2006:1-24.
- Talari ACS, Martinez MA, Movasaghi Z, et al. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl Spectrosc Rev 2017;52:456-506. [Crossref]
- Hashimoto E, Ishikawa M, Shinoda K, et al. Tissue classification of liver pathological tissue specimens image using spectral features. In: Gurcan MN, Tomaszewski JE, editors. Proc. SPIE 10140, Medical Imaging 2017: Digital Pathology. International Society for Optics and Photonics, Orlando, 2017:101400Z-101400Z-6.
- Zhang Y, Chen Y, Yu Y, et al. Visible and near-infrared spectroscopy for distinguishing malignant tumor tissue from benign tumor and normal breast tissues in vitro. J Biomed Opt 2013;18:077003. [Crossref] [PubMed]
- Coe JV, Chen Z, Li R, et al. Imaging infrared spectroscopy for fixation-free liver tumor detection. In: Farkas DL, Nicolau DV, Leif RC, editors. Proc. SPIE 8947, Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XII. International Society for Optics and Photonics, SPIE, San Francisco, 2014:89470B.
- Sheng D, Xu F, Yu Q, et al. A study of structural differences between liver cancer cells and normal liver cells using FTIR spectroscopy. J Mol Struct 2015;1099:18-23. [Crossref]
- Deshmukh SK, Srivastava SK, Poosarla T, et al. Inflammation, immunosuppressive microenvironment and breast cancer: opportunities for cancer prevention and therapy. Ann Transl Med 2019;7:593. [Crossref] [PubMed]
- Childs DTD, Hogg RA, Revin DG, et al. Sensitivity advantage of QCL tunable-laser mid-infrared spectroscopy over FTIR spectroscopy. Appl Spectrosc Rev 2015;50:822-39. [Crossref]
- Ukkonen H, Kumar S, Mikkonen J, et al. Changes in the microenvironment of invading melanoma and carcinoma cells identified by FTIR imaging. Vib Spectrosc 2015;79:24-30. [Crossref]
- Le TT, Rehrer CW, Huff TB, et al. Nonlinear optical imaging to evaluate the impact of obesity on mammary gland and tumor stroma. Mol Imaging 2007;6:205-11. [Crossref] [PubMed]
- Kiyotoki S, Nishikawa J, Okamoto T, et al. New method for detection of gastric cancer by hyperspectral imaging: a pilot study. J Biomed Opt 2013;18:26010. [Crossref] [PubMed]
- Goto A, Nishikawa J, Kiyotoki S, et al. Use of hyperspectral imaging technology to develop a diagnostic support system for gastric cancer. J Biomed Opt 2015;20:016017. [Crossref] [PubMed]
- Akbari H, Uto K, Kosugi Y, et al. Cancer detection using infrared hyperspectral imaging. Cancer Sci 2011;102:852-7. [Crossref] [PubMed]
- Siqueira LFS, Lima KMG. A decade (2004–2014) of FTIR prostate cancer spectroscopy studies: an overview of recent advancements. TrAC-Trend Anal Chem 2016;82:208-21. [Crossref]
- Pilling MJ, Henderson A, Shanks JH, et al. Infrared spectral histopathology using haematoxylin and eosin (H&E) stained glass slides: a major step forward towards clinical translation. Analyst 2017;142:1258-68. [Crossref] [PubMed]
- Sharma H, Zerbe N, Lohmann S, et al. A review of graph-based methods for image analysis in digital histopathology. Diagnostic pathology 2015;
- Großerueschkamp F, Kallenbach-Thieltges A, Behrens T, et al. Marker-free automated histopathological annotation of lung tumour subtypes by FTIR imaging. Analyst 2015;140:2114-20. [Crossref] [PubMed]
- Akalin A, Mu X, Kon MA, et al. Classification of malignant and benign tumors of the lung by infrared spectral histopathology (SHP). Lab Invest 2015;95:406-21. [Crossref] [PubMed]
- Mu X, Kon M, Ergin A, et al. Statistical analysis of a lung cancer spectral histopathology (SHP) data set. Analyst 2015;140:2449-64. [Crossref] [PubMed]
- Wang C, Zheng W, Bu Y, et al. Multi-scale hyperspectral imaging of cervical neoplasia. Arch Gynecol Obstet 2016;293:1309-17. [Crossref] [PubMed]
- Kallenbach-Thieltges A, Großerüschkamp F, Mosig A, et al. Immunohistochemistry, histopathology and infrared spectral histopathology of colon cancer tissue sections. J Biophotonics 2013;6:88-100. [Crossref] [PubMed]
- Beaulieu RJ, Goldstein SD, Singh J, et al. Automated diagnosis of colon cancer using hyperspectral sensing. Int J Med Robot 2018;14:e1897. [Crossref] [PubMed]
- Diem M, Ergin A, Mu X. Spectral histopathology of the lung: A review of two large studies. J Biophotonics 2019;12:e201900061. [Crossref] [PubMed]
- Akalin A, Ergin A, Remiszewski S, et al. Resolving Interobserver Discrepancies in Lung Cancer Diagnoses by Spectral Histopathology. Arch Pathol Lab Med 2019;143:157-73. [Crossref] [PubMed]
- Ly E, Piot O, Wolthuis R, et al. Combination of FTIR spectral imaging and chemometrics for tumour detection from paraffin-embedded biopsies. Analyst 2008;133:197-205. [Crossref] [PubMed]
- Urboniene V, Pucetaite M, Jankevicius F, et al. Identification of kidney tumor tissue by infrared spectroscopy of extracellular matrix. J Biomed Opt 2014;19:087005. [Crossref] [PubMed]
- Ali M H, Al-Saad K, Popelka A, et al. Application of Fourier transform infrared (FTIR) spectroscopy and atomic force microscopy in stroke-affected brain tissue. Swift J Med Med Sci 2016;2:011-024.
- Bergner N, Romeike BF, Reichart R, et al. Tumor margin identification and prediction of the primary tumor from brain metastases using FTIR imaging and support vector machines. Analyst 2013;138:3983-90. [Crossref] [PubMed]
- Wald N, Goormaghtigh E. Infrared imaging of primary melanomas reveals hints of regional and distant metastases. Analyst 2015;140:2144-55. [Crossref] [PubMed]
- Diem M, Miljković M, Bird B, et al. Cancer screening via infrared spectral cytopathology (SCP): results for the upper respiratory and digestive tracts. Analyst 2016;141:416-28. [Crossref] [PubMed]
- Menzies GE, Fox HR, Marnane C, et al. Fourier transform infrared for noninvasive optical diagnosis of oral, oropharyngeal, and laryngeal cancer. Transl Res 2014;163:19-26. [Crossref] [PubMed]
- Severcan F, Ozek NS, Gok S. Fourier Transform Infrared Spectroscopy and Imaging in Cancer Diagnosis and Characterization. Biophys J 2015;108:479a-80a. [Crossref]
- Zheng W, Wang C, Chang S, et al. Hyperspectral wide gap second derivative analysis for in vivo detection of cervical intraepithelial neoplasia. J Biomed Opt 2015;20:121303. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Neittaanmäki N, Salmivuori M, Pölönen I, et al. Hyperspectral imaging in detecting dermal invasion in lentigo maligna melanoma. Br J Dermatol 2017;177:1742-4. [Crossref] [PubMed]
- Bratchenko IA, Myakinin OO, Sherendak VP, et al. In vivo hyperspectral analysis of skin hemoglobin and melanin content for neoplasia detection. J-BPE 2018. doi:
10.18287/JBPE18.04.040301 . - Bratchenko IA, Sherendak VP, Myakinin OO, et al. In vivo hyperspectral imaging of skin malignant and benign tumors in visible spectrum. J-BPE 2018. doi:
10.18287/JBPE17.04.010301 . - Monheit G, Cognetta AB, Ferris L, et al. The performance of MelaFind: a prospective multicenter study. Arch Dermatol 2011;147:188-94. [Crossref] [PubMed]
- Fink C, Jaeger C, Jaeger K, et al. Diagnostic performance of the MelaFind device in a real-life clinical setting. J Dtsch Dermatol Ges 2017;15:414-9.91.
- Kim S, Cho D, Kim J, et al. Smartphone-based multispectral imaging: system development and potential for mobile skin diagnosis. Biomed Opt Express 2016;7:5294-307. [Crossref] [PubMed]
- Halicek M, Lu G, Little JV, et al. Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J Biomed Opt 2017;22:60503. [Crossref] [PubMed]
- Lu G, Wang D, Qin X, et al. Detection and delineation of squamous neoplasia with hyperspectral imaging in a mouse model of tongue carcinogenesis. J Biophotonics 2018;11: [Crossref] [PubMed]
- Bedard N, Schwarz RA, Hu A, et al. Multimodal snapshot spectral imaging for oral cancer diagnostics: a pilot study. Biomed Opt Express 2013;4:938-49. [Crossref] [PubMed]
- Gu X, Han Z, Yao L, et al. Image enhancement based on in vivo hyperspectral gastroscopic images: a case study. J Biomed Opt 2016;21:101412. [Crossref] [PubMed]
- Lim HT, Murukeshan VM. A four-dimensional snapshot hyperspectral video-endoscope for bio-imaging applications. Sci Rep 2016;6:24044. [Crossref] [PubMed]
- Hohmann M, Kanawade R, Klämpfl F, et al. In-vivo multispectral video endoscopy towards in-vivo hyperspectral video endoscopy. J Biophotonics 2017;10:553-64. [Crossref] [PubMed]
- Sullivan R, Alatise OI, Anderson BO, et al. Global cancer surgery: delivering safe, affordable, and timely cancer surgery. Lancet Oncol 2015;16:1193-224. [Crossref] [PubMed]
- Shapey J, Xie Y, Nabavi E, et al. Intraoperative multispectral and hyperspectral label-free imaging: A systematic review of in vivo clinical studies. J Biophotonics 2019;12:e201800455. [Crossref] [PubMed]
- Schols RM, Connell NJ, Stassen LP. Near-infrared fluorescence imaging for real-time intraoperative anatomical guidance in minimally invasive surgery: a systematic review of the literature. World J Surg 2015;39:1069-79. [Crossref] [PubMed]
- Sorg BS, Moeller BJ, Donovan O, et al. Hyperspectral imaging of hemoglobin saturation in tumor microvasculature and tumor hypoxia development. J Biomed Opt 2005;10:44004. [Crossref] [PubMed]
- Zuzak KJ, Cadeddu JA, Ufret-vincenty R, et al. Digital light processing hyperspectral imaging apparatus and method. United States Patent 9622662, 2017.
- Holzer MS, Best SL, Jackson N, et al. Assessment of renal oxygenation during partial nephrectomy using hyperspectral imaging. J Urol 2011;186:400-4. [Crossref] [PubMed]
- Tetschke F, Markgraf W, Gransow M, et al. Hyperspectral imaging for monitoring oxygen saturation levels during normothermic kidney perfusion. J Sens Sens Syst 2016;5:313-8. [Crossref]
- Pichette J, Laurence A, Angulo L, et al. Intraoperative video-rate hemodynamic response assessment in human cortex using snapshot hyperspectral optical imaging. Neurophotonics 2016;3:045003. [Crossref] [PubMed]
- Baltussen EJM, Kok END, Brouwer de Koning SG, et al. Hyperspectral imaging for tissue classification, a way toward smart laparoscopic colorectal surgery. J Biomed Opt 2019;241-9. [PubMed]
- Zuzak KJ, Naik SC, Alexandrakis G, et al. Characterization of a near-infrared laparoscopic hyperspectral imaging system for minimally invasive surgery. Anal Chem 2007;79:4709-15. [Crossref] [PubMed]
- Lu G, Little JV, Wang X, et al. Detection of Head and Neck Cancer in Surgical Specimens Using Quantitative Hyperspectral Imaging. Clin Cancer Res 2017;23:5426-36. [Crossref] [PubMed]
- Kho E, de Boer LL, Van de Vijver KK, et al. Hyperspectral Imaging for Resection Margin Assessment during Cancer Surgery. Clin Cancer Res 2019;25:3572-80. [Crossref] [PubMed]
- Boppart SA, Brown JQ, Farah CS, et al. Label-free optical imaging technologies for rapid translation and use during intraoperative surgical and tumor margin assessment. J Biomed Opt 2017;23:1-10. [Crossref] [PubMed]
- Halicek M, Little JV, Wang X, et al. Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks. J Biomed Opt 2019;24:1-9. [Crossref] [PubMed]
- Kho E, Dashtbozorg B, de Boer LL, et al. Broadband hyperspectral imaging for breast tumor detection using spectral and spatial information. Biomed Opt Express 2019;10:4496-515. [Crossref] [PubMed]
- Fabelo H, Ortega S, Ravi D, et al. Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations. PLoS One 2018;13:e0193721. [Crossref] [PubMed]
- Fabelo H, Ortega S, Lazcano R, et al. An Intraoperative Visualization System Using Hyperspectral Imaging to Aid in Brain Tumor Delineation. Sensors (Basel) 2018;18:430. [Crossref] [PubMed]
- Fabelo H, Ortega S, Szolna A, et al. In-Vivo Hyperspectral Human Brain Image Database for Brain Cancer Detection. IEEE Access 2019;7:39098-116.
- Pike R, Lu G, Wang D, et al. A Minimum Spanning Forest-Based Method for Noninvasive Cancer Detection With Hyperspectral Imaging. IEEE Trans Biomed Eng 2016;63:653-63. [Crossref] [PubMed]
- Lu G, Wang D, Qin X, et al. Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery. J Biomed Opt 2015;20:126012. [Crossref] [PubMed]
- Sucher R, Athanasios A, Köhler H, et al. Hyperspectral Imaging (HSI) in anatomic left liver resection. Int J Surg Case Rep 2019;62:108-11. [Crossref] [PubMed]
- Fei B, Lu G, Wang X, et al. Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients. J Biomed Opt 2017;22:1-7. [Crossref] [PubMed]
- Bao J, Bawendi MG. A colloidal quantum dot spectrometer. Nature 2015;523:67-70. [Crossref] [PubMed]


