
The overexpression of ZWINT in integrated bioinformatics analysis forecasts poor prognosis in breast cancer
Introduction
Breast cancer is a malignant tumor originating from the mammary epithelial tissue. It is the leading cause of cancer deaths among women and the disease with the highest incidence of cancer among women in the world (1). Therefore, in-depth study of the pathogenesis of breast cancer, the search for potential therapeutic targets and prognostic evaluation of biomarkers has become a research hotspot in this field. Numerous studies reported that the Zw10 binding factor (Zeste White 10 interactor, ZWINT) encoded by the ZWINT gene is a protein that regulates centromere division. It is a key regulatory protein in mitotic checkpoints and could regulate the cell cycle (2). The cell cycle checkpoint could also regulate the cell cycle. When the cell cycle runs to the checkpoint, it will be tested. The previous phase is completed before entering the next phase (3,4). In addition, it has been reported that ZWINT is associated with chromosome instability (CIN), and the abnormal number of chromosomes caused by CIN is considered to be a marker for a variety of human malignancies (5). Therefore, we speculate that ZWINT and tumor development and development are closely related. Although it has been reported that ZWINT is expressed in many tumors [such as ovarian cancer (6), liver cancer (7)], little research has been done on its expression in breast cancer.
Oncomine is the world’s largest cancer gene chip database, which is also an integrated data mining platform. It has collected 715 gene expression data sets, 86,733 cancer tissues and normal tissue samples. The integrated literature and chip data of this platform are obtained due to high quality. Highly recognized by researchers. The Kaplan-Meier Plotter database is currently an extensive online database for prognosis analysis, covering more than 5,100 breast cancer samples, with a prognostic analysis of nearly 55 thousand genes and more objective results.
Oncomine database and Kaplan-Meier Plotter database were used in this study to delve into the ZWINT’s expression in BC and its impact on the prognosis, which provided clues for further study on the mechanism of breast cancer development.
Methods
Oncomine database analysis
The expression of ZWINT gene in different types of cancers is defined in the Oncomine database (https://www.oncomine.org/resource/login.html) (8). The threshold is determined based on the following values, with the P value of 0.001, the fold changes of 2, and genes ranking of all.
Kaplan-Meier plotter database analysis
Using 10,461 cancer samples, the Kaplan-Meier plotter was able to assess the effect of 54,675 genes on survival. These cancer samples consisted of 5,143 breast cancer, 1,816 ovarian cancer, 2,437 lung cancer and 1,065 gastric cancer, which were located on the HGU133 Plus 2.0 array, respectively. The mean follow-up time of these cancer samples was 69, 40, 49 and 33 months, respectively. Kaplan-Meier plotter was also used to analyze the relationship between ZWINT expression and survival rates of breast cancer, ovarian cancer, lung cancer and gastric cancer (http://kmplot.com/analysis/) (9). The hazard ratio (HR) of 95% confidence interval (CI) and logarithmic rank P was calculated.
Approval was waived by the local ethics committee, as Oncomine database and Kaplan-Meier plotter database are publicly available and de-identified.
Statistical analysis
Survival curve is generated by Kaplan-Meier plots. The results generated in Oncomine showed P values, fold changes and grades. The results of Kaplan-Meier plot showed that HR and P<0.05 were considered to be statistically significant.
Results
The mRNA expression of ZWINT in various human cancers
To determine the difference of ZWINT expression between tumors and normal tissues, the oncogenic amine database was used to analyze the expression level of ZWINT mRNA in different tumors and normal tissues of different types of tumors. The expression of ZWINT in breast cancer, lung cancer, sarcoma, ovarian cancer, bladder cancer, liver cancer and cervical cancer was higher than that in normal tissues (Figure 1). In addition, the expression of ZWINT was lower in gastric cancer, prostate cancer, myeloma, renal cancer and pancreatic cancer in certain data sets. In the database, ZWINT gene was highly expressed in 75 studies. Fourteen studies were related to breast cancer.
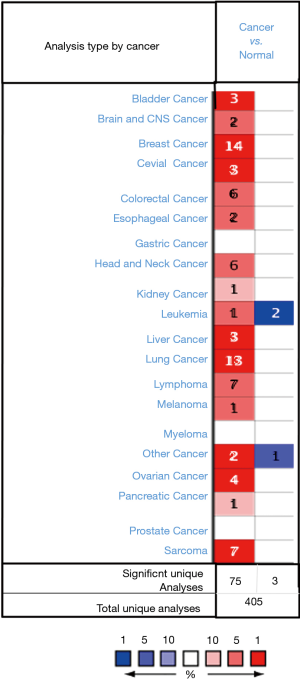
Expression of ZWINT in breast cancer and normal breast tissue
ONCOMINE analysis demonstrated that the expression of ZWINT in breast cancer was significantly higher than that in normal cells. In one set of data, ZWINT transcripts in 137 samples of TCGA (Cancer Genome Mapping) database (Figure 2A) increased by 4.133 times compared with normal tissues. In the study of Zhao (10), ZWINT in breast cancer samples increased by 2.313f (P=1.09e−8) compared with normal tissue (Figure 2B).

By meta-analysis of 22 studies in oncology database, ZWINT gene ranked 412 out of all differentially expressed genes, showing significant overexpression in breast cancer tissues compared with normal tissues (P=4.05E−6) (Figure 2C). The results of these 22 studies were published in the journals such as Mol Biol Cell (10), Proc Natl Acad Sci USA (11), Nature (12,13), Breast Cancer Res (14), BMC Cancer (15), Cancer Cell (16), Breast Cancer Res Treat (17), and Nat Med (18).
Prognostic value in ZWINT breast cancer
We examined whether the expression of ZWINT was related to the prognosis of breast cancer patients. The effect of the ZWINT representation on the survival rate was evaluated using the Kaplan-Meier plotter database. Note that the expression of ZWINT significantly affects the prognosis of breast cancer. As a result, in the case of all BC patients, the high expression of the ZWINT mRNA was related to worse overall survival (OS) (HR =1.73, 95% CI: 1.39–2.51, P=5.4×10−7), RFS (HR =1.68, 95% CI: 1.51–1.88, P<1×10−16) and distant metastasis-free survival (DMFS) (HR =1.55, 95% CI: 1.28–1.89, P=7.9×10−6) in all BC patients (Figure 3). Therefore, the ZWINT high expression is an independent risk factor and is thought to cause poor prognosis in patients with BC.
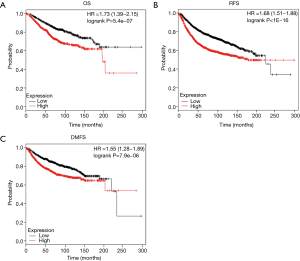
Discussion
According to the latest report of the American Cancer Association, the incidence of female breast cancer in 2007–2013 has risen. From 1989 to 2015, the death rate of breast cancer in America has fallen by about 39 percent (19). According to the study, this may be due to the development of human epidermal growth factor receptor 2, vascular endothelial growth factor and epidermal growth factor receptor and the application of targeted drugs to the treatment of breast cancer (20-22). However, since these target points exist only in some breast cancer patients, it is important to develop key targets for breast cancer development and to develop new target drugs for breast cancer treatment.
The ZWINT protein, encoded by the ZWINT gene, is a protein that regulates centromere division. It binds to Zw10 and colocalizes on the centromere and attaches to the microtubules of chromosomes and spindles. It is a chromosomal movement and mitotic checkpoint, which is an important regulatory protein that is associated with chromosomal instability (CIN). The abnormal number of chromosomes caused by CIN is considered to be a marker for many human malignancies (13). ZWINT has been reported to be overexpressed in different human cancers, but there are fewer studies in breast cancer.
This study employed independent data sets from the Oncomine database and Kaplan-Meier plotter databases to detect the expression levels of ZWINT and prognostic landscape in breast cancer. In this study, differential expression of ZWINT was observed between tumors and normal tissues. Based on the Oncomine database, we found that the expression of ZWINT in breast cancer, lung cancer, sarcoma, ovarian cancer, bladder cancer, liver cancer and cervical cancer was higher than that in normal tissues, while other data sets showed that the expression of ZWINT was lower in gastric, prostate, myeloma, kidney and pancreatic cancers (Figure 1). By further analyzing the expression of ZWINT in breast cancer and normal breast tissue, we found that the expression of ZWINT in breast cancer was significantly higher than that in normal breast tissue. In addition, meta-analysis showed that ZWINT gene ranked 412 out of all differentially expressed genes, and its expression in breast cancer tissues was much higher than that in normal tissues (P=4.05×10−6) (Figure 2C). In addition, data analysis by Kaplan-Meier plotter showed that high expression of ZWINT was associated with high HR of OS, recurrence-free survival (RFS) and DMFS (Figure 3). The reason may be that breast cancer occurs in vivo, making the expression of ZWINT gene increase, which in turn increases the growth, migration or invasion of tumors. Consequently, this is not conducive to the prognosis of patients (23), but its specific expression in vivo is significantly increased. In conclusion, these findings suggest that high expression of ZWINT is associated with poor prognosis of breast cancer, and ZWINT can be a biomarker of prognosis of breast cancer. Peng et al. (24) indicated that Knockdown of ZWINT inhibited cell behavior and growth. Meanwhile, ZWINT knockdown also retrained tumor volume in vivo. They think ZWINT may be a novel target for lung cancer therapy. However, whether ZWINT is a new target for breast cancer treatment requires further study.
In conclusion, the expression of ZWINT in breast cancer was significantly higher than that in normal control group, and the survival rate of breast cancer patients was lower. ZWINT can be considered as a specific biomarker and an important prognostic factor for breast cancer.
Acknowledgments
Funding: This work was partially supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.66). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived by the local ethics committee, as Oncomine database and Kaplan-Meier plotter database are publicly available and de-identified.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dou Z, Prifti DK, Gui P, et al. Recent Progress on the Localization of the Spindle Assembly Checkpoint Machinery to Kinetochores. Cells 2019;8:278. [Crossref] [PubMed]
- Gorgoulis VG, Pefani DE, Pateras IS, et al. Integrating the DNA damage and protein stress responses during cancer development and treatment. J Pathol 2018;246:12-40. [Crossref] [PubMed]
- Lindström MS, Jurada D, Bursac S, et al. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018;37:2351-66. [Crossref] [PubMed]
- Vargas-Rondón N, Villegas VE, Rondon-Lagos M, et al. The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 2017;10:4. [Crossref] [PubMed]
- Xu Z, Zhou Y, Cao Y, et al. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis. Med Oncol 2016;33:130. [Crossref] [PubMed]
- Yang XY, Wu B, Ma SL, et al. Decreased Expression of ZWINT is Associated With Poor Prognosis in Patients With HCC After Surgery. Technol Cancer Res Treat 2018;17:1533033818794190. [Crossref] [PubMed]
- Pan JH, Zhou H, Cooper L, et al. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front Immunol 2019;10:6. [Crossref] [PubMed]
- Lánczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160:439-46. [Crossref] [PubMed]
- Zhao H, Langerod A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell 2004;15:2523-36. [Crossref] [PubMed]
- Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is over-expressed in breast cancer. Proc Natl Acad Sci USA 2005;102:11005-10. [Crossref] [PubMed]
- Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2000 breast tumors reveals novel subgroups. Nature 2012;486:346-52. [Crossref] [PubMed]
- Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumor stroma promote breast cancer metastasis. Nature 2007;449:557-63. [Crossref] [PubMed]
- Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009;11:R7. [Crossref] [PubMed]
- Turashvili G, Bouchal J, Baumforth K, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer 2007;7:55. [Crossref] [PubMed]
- Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006;9:121-32. [Crossref] [PubMed]
- Glück S, Ross JS, Royce M, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/- trastuzumab. Breast Cancer Res Treat 2012;132:781-91. [Crossref] [PubMed]
- Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008;14:518-27. [Crossref] [PubMed]
- DeSantis CE, Ma J, Goding SA, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017;67:439-48. [Crossref] [PubMed]
- Mendes D, Alves C, Afonso N, et al. The benefit of HER2-targeted therapies on overall survival of patients with metastatic HER2-positive breast cancer-a systematic review. Breast Cancer Res 2015;17:140. [Crossref] [PubMed]
- Mayer IA, Abramson VG, Formisano L, et al. A Phase Ib study of alpelisib (BYL719), a PI3 Kα-specific inhibitor, with letrozole in ER+/HER2-metastatic breast cancer. Clin Cancer Res 2017;23:26. [Crossref] [PubMed]
- Baselga J, Gómez P, Greil R, et al. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol 2013;31:2586-92. [Crossref] [PubMed]
- Ying H, Xu ZY, Chen MM, et al. Overexpression of Zwint predicts poor prognosis and promotes the proliferation of hepatocellular carcinoma by regulating cell-cycle-related proteins. Onco Targets Ther 2018;11:689-702. [Crossref] [PubMed]
- Peng F, Li Q, Niu SQ, et al. ZWINT is the next potential target for lung cancer therapy. J Cancer Res Clin Oncol 2019;145:661-73. [Crossref] [PubMed]


