
The analysis of a ceRNA network and the correlation between lncRNA, miRNA, and mRNA in bladder cancer
Introduction
Bladder cancer is a malignant tumor originating from the bladder uroepithelium. It is one of the most common malignant tumors in the urinary system and seriously endangers human health. The incidence of bladder cancer in the world ranks 11th among all malignant tumors, among which the incidence rate in males is 9.0/100,000, ranking 7th among male malignant tumors. In China, the incidence of bladder cancer in men has exceeded that of prostate cancer, ranking first among malignant tumors in the male urinary system (1) (Standard for Diagnosis and Treatment of Bladder Cancer, 2018 Edition).
In recent years, great progress has been made in the study of bladder cancer, but its mechanisms are still unclear. Studies have shown that gene mutation and abnormal regulation of gene expression play an important role in the process of tumorigenesis and progression, and also play an important role in the occurrence and development of bladder cancer (2,3). MicroRNA (miRNA), for example, has been widely shown to be involved in the development of bladder cancer (4). Compared with miRNAs, there are few studies on newly discovered long non-coding RNA (lncRNA). lncRNA is a non-coding RNA transcript with a length of more than 200 bp, which does not encode proteins. LncRNA was once thought to have no biological functions and to be a “noise” in transcription. However, recent studies have shown that lncRNA plays an important role in epigenetic regulation, transcriptional regulation, and post-transcriptional regulation by acting as a signaling molecule, bait molecule, scaffold molecule, or guide molecule in the form of RNA, and participates in nearly all physiological and pathological processes of organisms. The abnormal expression of lncRNAs is closely related to the occurrence, development, and metastasis of tumors (5,6). The effect of abnormal expression of lncRNA in bladder cancer tissues on the proliferation, invasion, and migration of bladder cancer cells, and its guiding significance for clinical diagnosis and prognosis have been reported (7,8). miRNA is a non-coding single-stranded RNA encoded by endogenous genes. Its length is 22 bp. It can regulate the expression of mRNA through the microRNA response element (MRE) on the mRNA (9). Each miRNA can regulate the transcription and translation of multiple RNA, and each MRE can interact with multiple miRNAs (10,11).
Salmena et al. (12) proposed the endogenous competitive non-coding ceRNA hypothesis, indicating that RNA transcripts can communicate with each other through microRNA response elements (MRES), thus affecting the expression level of various RNAs, which plays a crucial role in tumorigenesis. Although progress has been made in the study of bladder cancer, the detection of specific lncRNAs associated with bladder cancer, survival analysis, and other clinically significant lncRNAs have not been reported. Therefore, we attempted to extract sample data from the Cancer Genome Atlas (TCGA) database, conduct differential expression, GO, KEGG, and survival analysis. The aim of our study was to analyze the molecular functions of the integrated lncRNA itself and the related signaling pathways, and to find the related regulatory molecules. We hope our findings can provide a reliable theoretical basis for exploring the molecular mechanism of bladder cancer and finding molecular targets for clinical diagnosis or treatment.
Methods
Data acquisition of bladder cancer and its adjacent samples
All the data in this study were collected from the TCGA database until January 28, 2016, and included 408 cases of expression of mRNA, 407 cases of expression of miRNAs, and 412 cases of clinical samples, along with 19 matched samples of cancer tissues and adjacent normal tissues. After eliminating the duplicate data and ultra-low expression data, only part of the data information and incomplete clinical information were recorded in the follow-up research.
Screening of lncRNAs, miRNAs, and mRNAs differentially expressed and significantly surviving in bladder cancer tissues
The data of RNA expression profiles were collected, and the differentially expressed lncRNAs, miRNAs, and RNAs were analyzed by R language edgeR package. The differentially expressed lncRNAs, miRNAs and mRNAs whose absolute value of fold change (FC) was more than 2, and whose false discovery rate (FDR) was less than 0.05, were screened. At the same time, the volcano maps were drawn by ggplot2 package, and the differentially expressed lncRNAs, miRNAs, and mRNA were obtained. Survival analysis was then conducted by using R language Survival package, and lncRNAs, miRNAs, and mRNAs with significant differences in expression and survival analysis were screened out.
Correlation analysis between lncRNA and miRNA-RNA, and construction of a ceRNA network in bladder cancer
The expression data of differentially expressed and surviving significant RNA, lncRNA, and miRNAs were correlated with corresponding expression data (i.e., differentially expressed and surviving significant RNA corresponding to lncRNA and miRNA), and the correlation coefficient between them was calculated. The absolute value of the correlation coefficient equal to or more than 0.5 represented a significant correlation.
The interacting miRNAs were matched according to the miR-code database (http://www.mircode.org/) using the differentially expressed and surviving significant lncRNA and miRNAs from the above analysis (13). Target Scan (http://www.targetscan.org/vert_72/), miRDB (http://mirdb.org/), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) databases were used to predict the target genes of miRNAs (14-16), and the target genes present in all three databases were screened. Finally, a subset of sections was obtained from the differentially expressed and surviving target genes screened, and the target genes differentially expressed and significantly surviving from earlier analysis. Based on the above-obtained lncRNA, miRNAs, and RNA, a ceRNA network between lncRNA and miRNAs was constructed using Cytoscape 3.7.1, and Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes Genomes (KEGG) enrichment analysis of the target genes in the network were carried out.
Enrichment analysis and survival analysis
The differentially expressed and surviving significant mRNAs, lncRNAs, significantly correlated mRNAs and miRNAs, along with significantly correlated mRNAs were analyzed by GO functional annotation and KEGG pathway enrichment, respectively. Combined with the survival data of patients with bladder cancer in TCGA, the K-M curve method was used to analyze the survival of patients with bladder cancer using the R language Survival package. The relationship between lncRNAs, miRNAs, and mRNAs and the prognosis of bladder cancer was analyzed. P<0.05 was considered to have statistical significance.
Results
Differentially expressed and significantly surviving lncRNAs in bladder cancer
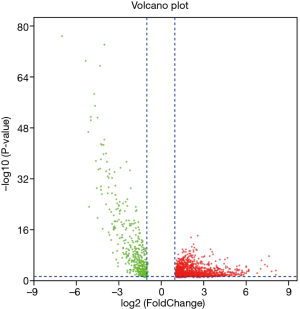
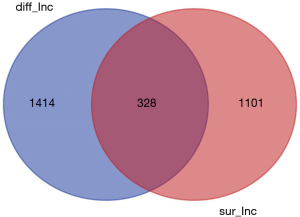
lncRNA was extracted from the expression matrix file downloaded from the TCGA database, and 1,742 differentially expressed lncRNAs were screened according to the criteria of |log FC| (>2), FDR <0.05 using R language edgeR package. As shown in the volcano map (Figure 1), there were 1,292 up-regulated lncRNAs and 450 down-regulated lncRNAs. Survival analysis of differentially expressed lncRNAs showed that 328 differentially expressed and significantly analyzed lncRNAs were screened. As shown in the Wayne diagram (Figure 2), 260 of them were up-regulated, and 68 were down-regulated.
Differentially expressed and significantly surviving miRNAs in bladder cancer
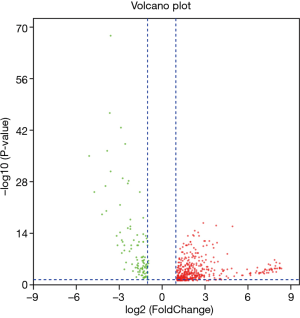
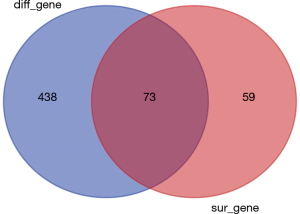
To study the direct relationship between lncRNA and miRNAs, the differentially expressed miRNAs of bladder cancer in the TCGA database were analyzed. In total, 511 differentially expressed miRNAs were identified according to the screening criteria of |log FC|(>2) and FDR <0.05, of which 406 were differentially up-regulated, and 105 were differentially down-regulated (Figure 3). By performing K-M survival analysis on differentially expressed miRNAs, a total of 73 differentially expressed and surviving miRNAs were obtained (Figure 4), of which 41 were up-regulated and 32 were down-regulated.
mRNA differentially expressed and significantly survived in bladder cancer
Similarly, according to the above method, the mRNA expression profiles of bladder cancer tissues and adjacent tissues in the TCGA database were compared, and differential mRNAs with |log FC| ≥2 and FDR <0.05 were screened. The results showed that there were 4,373 differentially expressed mRNAs in bladder cancer tumor tissues compared with normal tissues (Figure 5), of which 2,254 were up-regulated mRNA and 2,119 were down-regulated mRNA. The differentially expressed mRNA was subjected to survival analysis, and 677 mRNAs with significant differences and survival were screened (Figure 6), of which 246 were up-regulated and 431 were down-regulated.
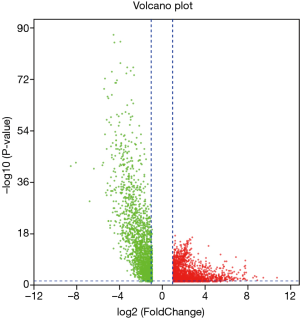
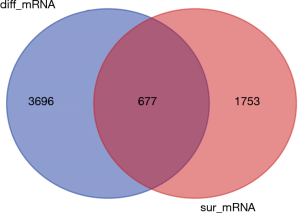
Construction of ceRNA network of lncRNA-miRNA-mRNA in bladder cancer
To further understand the role of differentially expressed lncRNAs in bladder cancer, we performed a correlation analysis between lncRNAs, miRNAs and mRNAs with variability of expression and significant survival. With 328 differentially expressed and significantly surviving lncRNAs and 73 miRNAs, the lncRNA-miRNA-mRNA ceRNA network was constructed. The results showed that 45 lncRNAs were involved in the regulation of 14 miRNAs (Table 1). After a subset of sections was obtained from 14 miRNA-regulated target genes and differentially expressed and surviving mRNAs, 29 target genes regulated by miRNAs were identified. Thus, 45 lncRNAs, 14 miRNAs, and 29 mRNAs constitute a direct regulatory relationship of lncRNA-miRNA-mRNA in bladder cancer (Figure 7).
Table 1
| miRNA | lncRNA | Q-value | R |
|---|---|---|---|
| hsa-miR-217-5p | SMC2-AS1 | 8.71E-262 | 0.974328 |
| hsa-miR-216a-5p | SMC2-AS1 | 6.01E-261 | 0.974076 |
| hsa-miR-204-5p | AC021242.3 | 3.32E-199 | 0.946584 |
| hsa-miR-192-5p | HNF1A-AS1 | 2.59E-141 | 0.893255 |
| hsa-miR-99a-5p | MIR99AHG | 5.49E-130 | 0.877411 |
| hsa-miR-99a-3p | MIR99AHG | 7.44E-106 | 0.834522 |
| hsa-let-7c-5p | MIR99AHG | 4.28E-101 | 0.824251 |
| hsa-let-7c-3p | MIR99AHG | 2.50E-88 | 0.792994 |
| hsa-miR-125b-2-3p | MIR99AHG | 2.18E-81 | 0.773362 |
| hsa-miR-504-5p | AC008537.3 | 2.26E-73 | 0.747935 |
| hsa-miR-217-5p | AC117489.1 | 2.48E-72 | 0.744398 |
| hsa-miR-216a-5p | AC117489.1 | 5.67E-72 | 0.74316 |
| hsa-miR-125b-5p | MIR99AHG | 1.25E-69 | 0.734916 |
| hsa-miR-100-5p | MIR100HG | 1.42E-53 | 0.668764 |
| hsa-miR-133b | ADAMTS9-AS2 | 1.70E-48 | 0.643551 |
| hsa-miR-133a-3p | ADAMTS9-AS2 | 3.79E-44 | 0.619897 |
| hsa-miR-143-3p | ADAMTS9-AS2 | 2.28E-42 | 0.609598 |
| hsa-miR-200a-3p | AL390719.2 | 2.13E-41 | 0.603821 |
| hsa-miR-145-3p | ADAMTS9-AS2 | 1.52E-40 | 0.598635 |
| hsa-miR-192-5p | SATB2-AS1 | 1.83E-39 | 0.591936 |
| hsa-miR-504-5p | LINC01152 | 3.75E-37 | 0.577061 |
| hsa-miR-133a-3p | CARMN | 3.00E-36 | 0.571033 |
| hsa-miR-323a-3p | VLDLR-AS1 | 5.78E-36 | 0.569108 |
| hsa-miR-431-5p | VLDLR-AS1 | 1.48E-35 | 0.566324 |
| hsa-miR-504-5p | AP001107.5 | 6.03E-35 | 0.562112 |
| hsa-miR-758-5p | VLDLR-AS1 | 6.35E-35 | 0.561957 |
| hsa-miR-377-5p | VLDLR-AS1 | 7.11E-35 | 0.561616 |
| hsa-miR-133b | AP000892.3 | 2.96E-34 | 0.557273 |
| hsa-miR-133a-3p | ADAMTS9-AS1 | 5.03E-34 | 0.555639 |
| hsa-miR-654-3p | VLDLR-AS1 | 6.21E-34 | 0.55499 |
| hsa-miR-133a-3p | AP000892.3 | 2.25E-33 | 0.550976 |
| hsa-miR-379-3p | VLDLR-AS1 | 2.73E-33 | 0.550372 |
| hsa-miR-133b | CARMN | 7.19E-33 | 0.547319 |
| hsa-miR-217-5p | AC120049.1 | 1.92E-32 | 0.544188 |
| hsa-miR-216a-5p | AC120049.1 | 4.07E-32 | 0.541762 |
| hsa-miR-145-3p | CARMN | 9.15E-32 | 0.539135 |
| hsa-miR-145-3p | AP000892.3 | 1.94E-31 | 0.536667 |
| hsa-miR-337-3p | VLDLR-AS1 | 3.14E-30 | 0.527373 |
| hsa-miR-192-5p | PTPRD-AS1 | 6.65E-30 | 0.524818 |
| hsa-miR-411-5p | VLDLR-AS1 | 1.29E-29 | 0.52254 |
| hsa-miR-143-3p | AP000892.3 | 2.22E-29 | 0.52066 |
| hsa-miR-504-5p | MAGI2-AS3 | 5.43E-29 | 0.517546 |
| hsa-miR-379-5p | VLDLR-AS1 | 1.64E-28 | 0.51365 |
| hsa-miR-133b | ADAMTS9-AS1 | 1.91E-28 | 0.513102 |
| hsa-miR-5579-3p | HNF1A-AS1 | 5.75E-28 | 0.509164 |
| hsa-miR-133b | LINC00702 | 8.72E-28 | 0.507662 |
| hsa-miR-143-3p | CARMN | 2.83E-27 | 0.50337 |
lncRNA, long non-coding RNA; miRNA, microRNA.
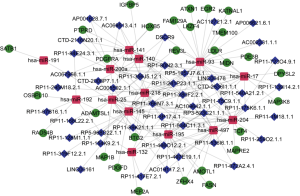
GO and KEGG enrichment analysis of mRNA in the lncRNA-miRNA-mRNA ceRNA network of bladder cancer
To further investigate the effect of mRNA in the relationship of lncRNA, miRN, and mRNA in bladder cancer on the cause and development of bladder cancer, GO and KEGG enrichment analysis was performed on lncRNA (differentially expressed and significantly surviving)-correlated mRNAs, miRNA (differentially expressed and significantly surviving)-correlated mRNAs, and differentially expressed and significantly surviving mRNAs. The results showed that some target genes were enriched in the signaling pathways, including cell adhesion molecules (CAMs), MAPK signaling pathway, and PI3K Akt signaling pathway. MAPK signaling pathway has been associated with promoting cell proliferation (17,18), while PI3K-Akt signaling pathway has been associated with tumor invasion and metastasis (19). Other pathways have been associated with cancer, adhesion, and abnormality in gene transcription (Figures 8-10).
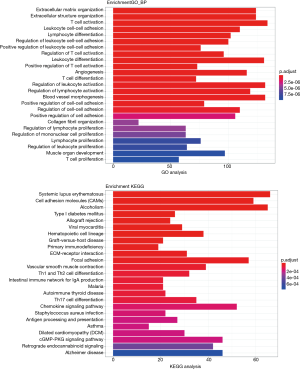

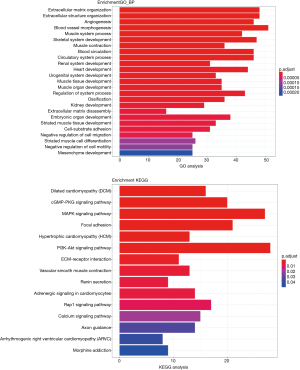
Similarly, we performed GO and KEGG enrichment analysis on mRNAs of the lncRNA-miRNA-mRNA ceRNA network, and the GO annotation results are shown in Figure 11 (only the top 10 are visible). The results of KEGG enrichment analysis showed that 29 mRNAs were involved in cancer-related signaling pathways such as fatty acid biosynthesis, gap junction, insulin signaling pathway, prostate cancer, regulation of actin cytoskeleton, and MAPK signaling pathway (Table 2).
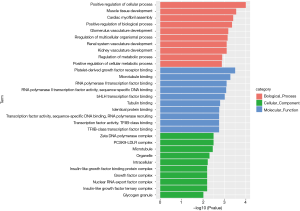
Table 2
| KEGGID | Pathway | Gene symbol | P value |
|---|---|---|---|
| 5218 | Melanoma | PDGFRA, PDGFD | 0.007544 |
| 61 | Fatty acid biosynthesis | FASN | 0.011313 |
| 4540 | Gap junction | PDGFRA, PDGFD | 0.011669 |
| 5215 | Prostate cancer | PDGFRA, PDGFD | 0.011669 |
| 4910 | Insulin signaling pathway | FASN, PDE3B | 0.026779 |
| 3440 | Homologous recombination | RAD54B | 0.051805 |
| 4510 | Focal adhesion | PDGFRA, PDGFD | 0.052877 |
| 4144 | Endocytosis | LDLR, PDGFRA | 0.053353 |
| 4810 | Regulation of actin cytoskeleton | PDGFRA, PDGFD | 0.058698 |
| 4010 | MAPK signaling pathway | MAP3K8, PDGFRA | 0.088056 |
KEGG, Kyoto Encyclopedia of Genes Genomes.
Discussion
Bladder cancer is the most common malignant tumor in the urinary system in China, and its incidence and mortality are the first in urinary tract tumors. Advances in medical technology have made great progress in the treatment of bladder cancer. However, the recurrence rate of bladder cancer is high, and the 5-year survival rate is low. Therefore, patients with bladder cancer need to endure long-term detection and treatment. The difficulties of this disease may be due to an insufficient understanding of the potential mechanism and the lack of effective biomarkers. Earlier studies have shown that improper gene regulation may play an important role in bladder cancer (20) and has enormous potential as a biomarker. This article studies the mechanism of action in bladder cancer to better understand and identify biomarkers of bladder cancer. With the development of high-throughput sequencing technology, more and more studies have shown that lncRNA plays a crucial role in the development of cancer (2,3), and that the expression of lncRNA is closely related to the progress of different cancers. Studies have shown that lncRNA TSLNC8 can interact with TKT and STAT3, thereby regulating the phosphorylation levels of STAT3-Tyr705 and STAT3-Ser727 and the transcriptional activity of STAT3, leading to the inactivation of the IL-6/STAT3 signaling pathway and suppressing the development of tumors (21). At present, the research focus of related lncRNA in urinary tract tumors at home and abroad is focused on prostate cancer. The related lncRNAs include prostate cancer gene expression marker 1 (PCGEM1), prostate-specific antigen 3 (DD3/PCA3), prostate non-coding RNA1 (PRNCR1), and others. In bladder tumors, there are more studies on urothelial carcinoma-associated gene 1 (UCA1) and imprinted gene H19, and more lncRNA studies are still in their infancy. In this study, a series of lncRNAs were screened using public data of RNA expression profiles from TCGA database. Target gene enrichment analysis revealed correlations with pathways such as the cGMP-PKG signaling pathway.
As an area of intense research, miRNA has been studied in relation to various tumors, including bladder cancer. It has been proven that miR-141, miR-200c, miR-30b, miR-370, miR-1236, and miRNAs like miR-144, miR-223, and miR-let-7, etc. (22-25) play a key role in the proliferation, invasion, metastasis, apoptosis, and prognosis evaluation of bladder cancer. Yonemori et al. (26) found that by down-regulating miR-139-5p or miR-139-3p to enhance the migration and invasion of bladder cancer cells, and directly target MMP11, which can be used as a prognostic marker for bladder cancer patients. In this study, bioinformatics methods were used to identify several miRNAs with differential expression and significant survival, such as MAP3K8 and PDGFRA, using the public resources of bladder cancer in TCGA database. Further enrichment analysis showed that these miRNA-related mRNAs were enriched in signaling pathways such as the MAPK signaling pathway, and this finding can provide a certain data foundation for further research.
The proposed ceRNA hypothesis provides an important guiding direction for the pathogenesis of tumors and also offers a new theoretical basis for the diagnosis and treatment of tumors. The interaction of RNA molecules like lncRNA, miRNA, and mRNA has important biological significance. Studies have shown that lncRNA, miRNA, and mRNA are in a state of equilibrium in the ceRNA network. Once this balanced microenvironment state is broken, cancer disease ensues (27). Studies have confirmed that lncRNA-regulated ceRNA networks have important regulatory effects in cancers such as gastric cancer, liver cancer, and prostate cancer (28-31). In this study, a ceRNA network consisting of 45 lncRNAs, 14 miRNAs, and 29 mRNAs was constructed by RNA omics data from TCGA database, which provided a basis for further experimental verification.
The inadequacy of this study is that all the data consist of changes in RNA levels provided by TCGA, with no data from the proteome, and thus we were unable to analyze mRNA at the protein level in ceRNA networks. In the next study, we will conduct quantitative analysis and experiments such as immunohistochemistry or Western blotting to further explore and verify the results of this study.
Conclusions
In summary, we performed a differential analysis of lncRNA, miRNA, and mRNA data in the TCGA database, and combined them with survival analysis to screen out those lncRNAs, miRNAs, and mRNAs that were differentially expressed and significantly proliferated. The correlation between lncRNA, miRNA, and mRNA was also analyzed in pairs, and a ceRNA network was constructed to provide a potential target for the diagnosis, treatment, and prognosis evaluation of bladder cancer.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.27). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Goebell PJ, Knowles MA. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol Oncol 2010;28:409-28. [Crossref] [PubMed]
- Rentsch CA, Müller DC, Ruiz C, et al. Comprehensive Molecular Characterization of Urothelial Bladder Carcinoma: A Step Closer to Clinical Translation? Eur Urol 2017;72:960-1. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013;34:613-20. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Martens-Uzunova ES, Böttcher R, Croce CM, et al. Long Noncoding RNA in Prostate, Bladder, and Kidney Cancer. Eur Urol 2014;65:1140-51. [Crossref] [PubMed]
- Zhang Q, Su M, Lu G, et al. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer 2013;12:101. [Crossref] [PubMed]
- Ye W, Lv Q, Wong CK, et al. The effect of central loops in miRNA: MRE duplexes on the efficiency of miRNA-mediated gene regulation. Plos One 2008;3:e1719. [Crossref] [PubMed]
- Amirkhah R, Schmitz U, Linnebacher M, et al. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer 2015;54:129-41. [Crossref] [PubMed]
- Wang C, Hu DZ, Liu JZ. Identification of critical TF-miRNA-mRNA regulation loops for colorectal cancer metastasis. Genet Mol Res 2015;14:5485-95. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Jeggari A, Marks DS, Larsson E. Larsson, miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012;28:2062-3. [Crossref] [PubMed]
- Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 2011;39:D163-169. [Crossref] [PubMed]
- Zhang C, Zhang CD, Ma MH, et al. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J Gastroenterol 2018;24:1206-15. [Crossref] [PubMed]
- Riffo-Campos ÁL, Riquelme I, Brebi-Mieville P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int J Mol Sci 2016;17:E1987. [Crossref] [PubMed]
- Hao YQ, Zheng HX. Study on the Mechanism of MAPK/ERK Signaling Pathway in Tumor Therapy. China Journal of Leprosy and Skin Diseases 2012;28:490-3.
- Chrzanowska-Wodnicka M. Regulation of angiogenesis by a small GTPase Rap1. Vascul Pharmacol 2010;53:1-10. [Crossref] [PubMed]
- Park JH, Kim JJ, Bae YS. Involvement of PI3K-AKT-mTOR pathway in protein kinase CKII inhibition-mediated senescence in human colon cancer cells. Biochem Biophys Res Commun 2013;433:420-5. [Crossref] [PubMed]
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15:25. [Crossref] [PubMed]
- Zhang J, Li Z, Liu L, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology 2018;67:171-87. [Crossref] [PubMed]
- Mahdavinezhad A, Mousavi-Bahar SH, Poorolajal J, et al. Evaluation of miR-141, miR-200c, miR-30b Expression and Clinicopathological Features of Bladder Cancer. Int J Mol Cell Med 2015;4:32-9. [PubMed]
- Wang C, Chen Z, Ge Q, et al. Up-regulation of p21WAF1/CIP1 by miRNAs and its implications in bladder cancer cells. FEBS Lett 2014;588:4654-64. [Crossref] [PubMed]
- Avgeris M, Mavridis K, Tokas T, et al. Uncovering the clinical utility of miR-143, miR-145 and miR-224 for predicting the survival of bladder cancer patients following treatment. Carcinogenesis 2015;36:528-37. [Crossref] [PubMed]
- Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 2009;125:345-52. [Crossref] [PubMed]
- Yonemori M, Seki N, Yoshino H, et al. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci 2016;107:1233-42. [Crossref] [PubMed]
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012;28:2062-3. [Crossref] [PubMed]
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. [Crossref] [PubMed]
- Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 2014;4:6088. [Crossref] [PubMed]
- Zhang R, Guo Y, Ma Z, et al. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget 2017;8:26079-89. [PubMed]
- Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13:92. [Crossref] [PubMed]


