
Proteomic and ultrastructural analysis of Clara cell and type II alveolar epithelial cell-type lung cancer cells
Introduction
Lung adenocarcinoma is the predominant histologic subtype of lung cancer in most countries. According to the histological heterogeneity found in clinical, imaging, molecular biological, and pathological studies, lung adenocarcinoma is divided into four subtypes: preinvasive lesions, minimally invasive adenocarcinoma, invasive adenocarcinoma, and variants of invasive adenocarcinoma (1). Lung adenocarcinoma cells can also be subclassified into six cytological subtypes by electron microscopy: bronchial cell surface type with little or no mucus, goblet cell type, bronchial gland cell type, Clara cell type, type II alveolar epithelial cell type, and mixed cell type (2-4).
However, adenocarcinomas of Clara cell and type II alveolar epithelial cell type are extremely similar under light microscopy and can only be identified by electron microscopy. Tumor cells of Clara cell type have a group of electron-dense secretory granules, an oval or round shape, and a diameter of 300–400 nm (Clara cell granules); whereas tumor cells of type II alveolar epithelial cell type have lamellar inclusion bodies with diameters of 400–600 nm within the cytoplasm (3). Therefore, Clara cell granules and lamellar inclusion bodies are the characteristic structures of Clara cell and type II alveolar epithelial cell-type lung adenocarcinoma cells, respectively. Problems associated with electron microscopy detection include a complicated tissue sampling process and a high cost of classification, requiring significant time and resources that are inconvenient for basic medical research and clinical applications.
Advanced “-omic” technologies, including proteomics, have opened up new avenues for gene expression profiling and detection of biomarkers for the identification of cytological subtypes of lung adenocarcinoma. In this study, we aimed to identify unique biomarkers for the molecular subtyping of Clara cell and type II alveolar epithelial cell-type adenocarcinomas with proteomic profiling. We then compared the molecular biomarker findings with the diagnosis rendered by electron microscopy.
Methods
Cells and antibodies
Six human lung adenocarcinoma cell lines (A549, NCI-H358, NCI-H1650, HCC827, NCI-H1395, and NCI-H1975) were purchased from the Cell Bank of the Chinese Academy of Sciences, China. Cells were cultured in RPMI 1640 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA). The primary antibodies used in the present experiments included mouse anti-human cytokeratin 19 (CK19) antibody (ZSGB-BIO, China), mouse anti-human cytokeratin 8 (CK8) antibody (ZSGB-BIO, China), rabbit anti-human peroxiredoxin 2 (PRDX2) antibody (Abcam, UK), mouse anti-human ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) antibody (Bioss, China), rabbit anti-human Clara cell 10-kD protein (CC10) (Santa Cruz, USA), rabbit anti-human pulmonary surfactant protein C (SP-C) (Proteintech, USA), and rabbit anti-human endoplasmic reticulum oxidoreductin 1-α (ERO1L) (Proteintech, USA). The present study does not need ethics approval because of no animals or people involved.
Electron microscopy
Cultured cells from the A549, NCI-H358, NCI-H1650, HCC827, NCI-H1395, and NCI-H1975 cell lines were digested with trypsin, washed with precooled phosphate-buffered saline (PBS), centrifuged at 1,000 × g and 4 °C for 5 min, and fixed with 2.5% glutaraldehyde at 4 °C overnight. After washing with PBS, the cells were fixed in 1% osmium tetroxide for 1 h and embedded in EPON resin. Sections of 50–70 nm in thickness were cut and stained with uranyl acetate/lead citrate and were then examined and photographed using a Philips CM-120 transmission electron microscope.
Protein sample preparation
Cultured A549 and NCI-H358 cells (passages 4–7) were collected and washed in precooled PBS, respectively. The total protein was extracted in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, pH 8.8) supplemented with protease and nuclease inhibitors (GE Healthcare, USA). The homogenate was centrifuged at 25,000 × g for 30 min, and the supernatants were collected. The protein samples were cleaned up with a clean-up kit and then quantified with a 2D-Quant kit (GE Healthcare, USA), according to the manufacturer’s protocols.
Two-dimensional difference gel electrophoresis (2D-DIGE) and image analysis
2D-DIGE was performed to identify differentially expressed proteins (DEPs) between A549 and NCI-H358 cells in four biological replicates (Table 1). Cy3 and Cy5 dye swap experiments were performed to avoid dye biasness. A 50-µg sample of cleaned-up proteins from A549 and NCI-H358 cells (passages 4–7), respectively, was mixed with 1.0 µL of diluted fluorescent dye (400 pmol), according to the manufacturer’s instructions. Samples from each cell line were respectively labeled with Cy3 or Cy5 (Abcam, UK), whereas a pooled sample by mixing equal amounts of protein from all samples in the experiment was labeled with Cy2 (Abcam, UK). Following labeling, the samples were incubated in the dark on ice for 30 min, and 1 µL of 10 mM lysine was added to terminate the reaction. Cy3-, Cy5-, and Cy2-labeled samples and internal standards were pooled and adjusted to equal volumes using rehydration buffer [8 M urea, 2% CHAPS, 0. 4% immobilized pH gradient (IPG) buffer, and 0.28% dithiothreitol (DTT)]. The samples were loaded onto an IPG gel strip (24 cm, linear, pH 4–7) for isoelectric focusing on an IPGphor isoelectric focusing system (GE Healthcare, USA). The parameters were 30 V for 6 h (rehydration) at 20 °C, 500 V for 1 h, gradient to 1,000 V for 1 h, gradient to 8,000 V for 1 h, and 8,000 V for 7 h. After isoelectric focusing, the strips were first equilibrated in equilibration solution of 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% (v/v) glycerol, 2% (w/v) sodium dodecyl sulfate, and 1% (w/v) DTT for 15 min, and then again in the same solution without DTT [DTT was replaced with 4% (w/v) iodoacetamide] for a further 15 min. The IPG strips were then loaded on the top of 12.5% polyacrylamide gels using an Ettan DALTsix system (GE Healthcare, USA) for electrophoresis at 2 W/gel overnight until the bromophenol blue tracking dye reached the bottom of the gels. The DIGE gels were scanned with a Typhoon 9400 laser scanner (GE Healthcare, USA) at a resolution of 100 µm to image each gel and detect the Cy2-, Cy3-, and Cy5-labeled proteins. The gel images were analyzed using DeCyder Differential in the Gel Analysis software (GE Healthcare, USA), according to the manufacturer’s protocols. The Student’s t-test was used to determine the DEPs between A549 and NCI-H358 cells. The spots that showed a statistically significant difference in abundance (variation of at least 4.5-fold, P<0.05) were used to generate a list of candidates for further analysis.
Table 1
| Gel No. | Cy3 | Cy5 |
|---|---|---|
| Gel 1 | A549 passage 4 | NCI-H358 passage 4 |
| Gel 2 | A549 passage 5 | NCI-H358 passage 5 |
| Gel 3 | NCI-H358 passage 6 | A549 passage 6 |
| Gel 4 | NCI-H358 passage 7 | A549 passage 7 |
2D-DIGE, two-dimensional difference gel electrophoresis.
In-gel digestion for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS/MS)
Two preparative gels were additionally made according to the same methods and conditions above (without fluorescent dye) and stained with PhastGel Blue R-250. Protein spots of interest were excised from the preparative gels with an Ettan Spot Picker (GE Healthcare, USA) and treated sequentially as follows: destaining, dehydration, desiccation, and then digestion with sequencing-grade trypsin solution (100 mM ammonium bicarbonate and 5 ng/µL trypsin) overnight at room temperature (5). The supernatant was transferred to a new Eppendorf tube, 100 µL of 60% acetonitrile/0.1% trifluoroacetic acid was added to the sediment, and the solution was ultrasonicated for 15 min. The supernatant was transferred and combined with the previous one and then freeze-dried.
The freeze-dried samples were dissolved in 2 µL of 20% acetonitrile for enzymatic hydrolysis. An aliquot of 1 µL was spotted directly onto the sample target plate. Once dried naturally, 0.5 µL of supersaturated α-cyano-4-hydroxycinnamic acid matrix solution in 50% acetonitrile and 0.1% trifluoroacetic acid was spotted onto the corresponding target plate and allowed to dry naturally. The sample target plate was cleaned with nitrogen and then placed in the target slot of the instrument for analysis with a tandem TOF-MS (4800 Plus MALDI TOF/TOFTM Analyzer). The laser source at a wavelength of 355 nm was from a Nd:YAG laser with an acceleration voltage of 2 kV. Data were collected using positive ion mode and automatic acquisition mode. The first-order MS scans ranged from 800 to 4,000 Da, whereas parent ions with a signal-to-noise ratio greater than 50 were selected and analyzed by secondary MS/MS. A total of 8 parent ions per sample point were selected with over 2,500 accumulation times by secondary MS/MS, 2 kV of collision energy, and closed collision-induced dissociation. Mascot 2.2 software was used to retrieve the original MS data with the following parameters: database, uniprot; taxonomy, Homo sapiens (133,549); type of search, combined (MS + MS/MS); enzyme, trypsin; fixed modifications, carbamidomethyl (c); dynamic modifications, oxidation (m); mass values, monoisotopic; protein mass, unrestricted; peptide mass tolerance, ± 100 ppm; fragment mass tolerance, ± 0.4 Da; peptide charge state, 1+; and maximum number of missed cleavages, 1.
2D western blot analysis
A 100-µg sample of cleaned-up protein from A549 or NCI-H358 cells was minimally labeled with Cy5 fluorescent dye, respectively. In the two directions of vertical electrophoresis, 1-cm-long filter paper containing a molecular weight marker was placed adjacent to the acidic end and the alkaline end of the adhesive strip. The remaining steps of the 2D-DIGE procedure were followed, without the inclusion of an internal standard. After electrophoresis, fluorescent images were captured by a Typhoon 9400 scanner (GE Healthcare, USA) at a resolution of 100 µm. A rectangular gel with proteins was excised from the whole gel to transfer proteins electrophoretically onto a polyvinylidene fluoride membrane (GE Healthcare, USA) with the following parameters: voltage, ≤25 V; current, total membrane area (cm2) × 0.8 (ma); and time of semi-dry electric transfer, 60 min. After transfer, the membrane was blocked with fluorescent special blocking solution for 2 h. After being washed briefly with PBS-Tween (PBS-T) buffer, the membrane was incubated overnight at 4 °C with rabbit anti-human CK19 antibody (1:50 dilution), mouse anti-human CK8 rabbit (1:50 dilution), rabbit anti-human ERO1L (1:100 dilution), rabbit anti-human PRDX2 antibody (1:250 dilution), or mouse anti-human UCHL1 antibody (1:100 dilution) diluted in PBS-T buffer. The membrane was washed extensively in PBS-T buffer for removal of the primary antibody, and then subsequently incubated with goat anti-mouse antibody (1:500) or goat anti-rabbit antibody (1:500) preadsorbed with Cy3 at room temperature for 2 h. The membrane was washed extensively in PBS-T buffer to remove the secondary antibody before it was imaged with a Typhoon 9400 scanner.
One-dimensional western blot analysis
Proteins extracted from A549, NCI-H358, NCI-H1650, HCC827, NCI-H1395, and NCI-H1975 cells were analyzed by one-dimensional western blot, according to standard protocols. Membranes were first incubated with the corresponding antibodies (ERO1L, 1:200 dilution; UCHL1, 1:300 dilution; CC10, 1:200 dilution; or SP-C, 1:200 dilution) or β-actin antibody (1:1,000 dilution) overnight at 4 °C, and then with secondary antibody conjugated with horseradish peroxidase for 1 h at room temperature. The intensity of the protein bands was quantified by densitometry and then analyzed using Image J software (NIH).
Statistical analysis
All data were expressed as the mean ± standard error of the mean and analyzed by the Student’s t-test and one-way analysis of variance. The least significant difference method was used for multiple comparisons between each group with SPSS 21.0 statistical software. The level of significance was set at P<0.05.
Results
Identification of six lung adenocarcinoma cell lines by electron microscopy
Ultrastructurally, six lung adenocarcinoma cell lines conformed with the ultrastructural characteristics of lung adenocarcinoma: large nuclei with an irregular folded membrane, prominent nucleoli, microvilli on the surface of the cells, microcavity in the cytoplasm, abundance of rough endoplasmic reticulum, and lysosomes.
The features of Clara cell differentiation were only observed in NCI-H358 cells, which showed dense secretory granules of 300–400 nm in diameter and a round or oval shape (Figure 1).
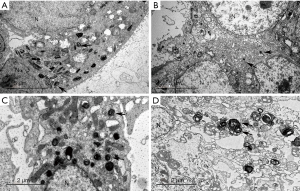
The features of type II alveolar epithelial cells were observed in A549, NCI-H1975, and HCC827 cells, which showed numerous lamellar inclusion bodies of 400–600 nm in diameter within the cytoplasm (Figure 1).
Differential characteristics of Clara cells or type II alveolar epithelial cells were not observed in NCI-H1395 or NCI-H1650 cells. None of the six cell lines showed characteristics of the bronchial surface cell type with little or no mucus production (e.g., tall columnar cells, the cytoplasm is rich in mitochondria and smooth-surfaced vesicles, a lack of secretory granules, cilia are very rarely observed, and basal bodies may exist), the goblet cell type (e.g., the cytoplasm is filled with mucous granules of rather low electron density, varying in internal structure), or the bronchial gland cell type (e.g., cuboidal or polygonal cells, mucous granules in the cytoplasm with various densities, and oval fibrillar structures) (3).
Screening and identification of DEPs by 2D-DIGE and MALDI-TOF-MS/MS
Protein samples isolated from four different generations of A549 (type II alveolar epithelial cell type) and NCI-H358 cells (Clara cell type) were separated by 2D-DIGE to produce images with Typhoon9400 (6-11). Miscellaneous spots were deleted manually, leaving 1275±196 remaining spots in each gel, from which a total of 735 spots were matched manually and automatically. Eight spots were differentially expressed by at least 4.5 times between the two cell lines (P<0.05), among which one spot (spot no. 1) was highly expressed in A549 cells, whereas seven spots (spot no. 2 to 8) were highly expressed in NCI-H358 cells. The ratio of the spot density and the P values of spots of each protein are shown in Table 2. The locations of the spots of each protein analyzed by 2D electrophoresis are shown in Figure 2. As shown in Figure 3, the expression of each of the eight spots was converted into three dimensions by DeCyder to facilitate analysis. Finally, seven interesting spots (spot no. 1, 2, 3, 5, 6, 7, and 8) were successfully detected in the preparative gels and excised for MALDI-TOF-MS/MS to identify the corresponding proteins. As shown in Table 3, spots 2, 3, and 5 are from the same protein. Therefore, a total of five proteins were identified successfully in the present study.
Table 2
| Spot No. | Ratio of spot density(A549/NCI-H358) | p |
|---|---|---|
| 1 | 11.06 | 0.00068 |
| 2 | −19.51 | 0.000048 |
| 3 | −15.07 | 0.000083 |
| 4 | −9.57 | 0.000069 |
| 5 | −9.17 | 0.0036 |
| 6 | −7.46 | 0.00029 |
| 7 | −5.22 | 0.0014 |
| 8 | −4.59 | 0.0042 |
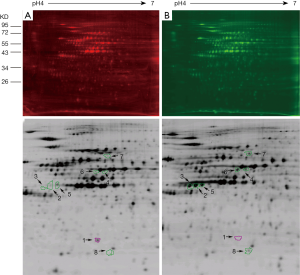
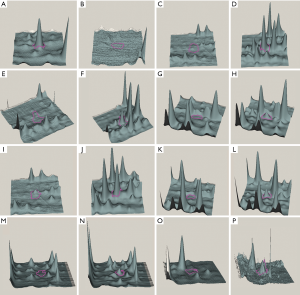
Table 3
| Spot No. | Name | MW (kDa)/PI | Accession | Score | Mascot score |
|---|---|---|---|---|---|
| 1 | UCHL1 | 25,150.6/5.33 | UCHL1_HUMAN | 307 | 205 |
| 2 | CK19 | 44,079.1/5.04 | K1C19_HUMAN | 533 | 395 |
| 3 | CK19 | 44,079.1/5.04 | K1C19_HUMAN | 819 | 552 |
| 5 | CK19 | 44,079.1/5.04 | K1C19_HUMAN | 679 | 408 |
| 6 | CK8 | 53,671.1/5.52 | K2C8_HUMAN | 815 | 554 |
| 7 | ERO1L | 55,213.4/5.48 | ERO1A_HUMAN | 697 | 471 |
| 8 | PRDX2 | 22,049.3/5.66 | PRDX2_HUMAN | 379 | 285 |
Seven spots (spot no. 1, 2, 3, 5, 6, 7, and 8) were successfully detected in the preparative gels and excised for MALDI-TOF-MS/MS to identify the corresponding proteins. Spot no. 4 was not detected in the preparative gels. UCHL1 was the only protein expressed at least 4.5 times higher in A549 cells than in NCI-H358 cells. CK19, CK8, ERO1L, and PRDX2 were expressed at least 4.5 times higher in NCI-H358 cells than in A549 cells. MALDI-TOF-MS/MS, matrix-assisted laser desorption/ionization; PI, isoelectric point; UCHL1, ubiquitin carboxyl-terminal hydrolase isozyme L1; ERO1L, endoplasmic reticulum oxidoreductin 1-α; CK, cytokeratin; PRDX2, peroxiredoxin 2.
Validation of DEPs with western blot
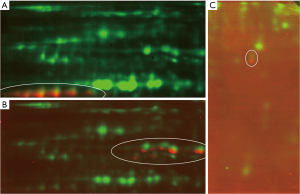
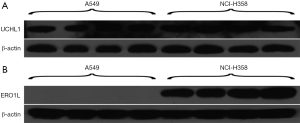
Western blot analysis was used to validate the expression levels of UCHL1, CK19, CK8, ERO1L, and PRDX2. As shown in Figure 4, CK19, CK8, and PRDX2 were successfully confirmed by 2D western blot. ERO1L and UCHL1 were further validated using one-dimensional western blot. The results showed a high expression of UCHL1 in both A549 and NCI-H358 cells, without a significant difference between the two cell lines (P=0.46). ERO1L was highly expressed in NCI-H358 cells, but no band was observed in A549 cells, showing a significant difference between the two cell lines (P=0.02) (Figure 5).
Protein expression levels of ERO1L, SP-C, and CC10 in NCI-H1650, HCC827, NCI-H1395, NCI-H1975, A549, and NCI-H358 cells
To further investigate whether ERO1L can be used as a promising biomarker to distinguish tumors of Clara cell and type II alveolar epithelial cell type, we used one-dimensional western blot to detect the expression levels of ERO1L as well as CC10 and SP-C in NCI-H1650, NCI-H1975, A549, NCI-H358, HCC827 and NCI-H1395 cells. The reason that we added CC10 and SP-C to the one-dimensional western blot validation was that CC10 exists in Clara granules of normal Clara cells (12-14), whereas SP-C is a human surfactant apoprotein in normal type II alveolar epithelial cells (15). It is assumed that CC10 and SP-C may be potential biomarkers for classification of Clara cell and type II alveolar epithelial cell-type lung adenocarcinoma cells. As shown in Figure 6, SP-C was expressed only in NCI-H1975 cells; ERO1L was expressed in NCI-H1650, NCI-H1975, HCC827, and NCI-H358 cells; and CC10 was expressed in all cell lines analyzed. ERO1L was expressed at a lower level in NCI-H1650 cells but at a higher level in NCI-H1975, HCC827, and NCI-H358 cells, showing statistically significant differences between pairs of NCI-H1650 cells with NCI-H1975, HCC827, and NCI-H358 cells (P<0.05).

Discussion
Clara cell and type II alveolar epithelial cell-type lung adenocarcinoma cells differ in terms of morphology under electron microscopy, but it still remains unclear how their biological behaviors differ and whether distinguishing them is meaningful for guiding clinical diagnosis and treatments. The development of appropriate biomarkers to replace tedious electron microscopic detection will greatly facilitate research on Clara cell and type II alveolar epithelial cell-type lung adenocarcinoma cells. In the present study, we used six lung adenocarcinoma cell lines to identify promising biomarkers for classification of the two cell types. Previous studies have demonstrated ultrastructural characteristics of type II alveolar epithelial cells in the NCI-H226, NCI-H726, NCI-H820, and NCI-H920 cell lines; Clara cell in the NCI-H358, NCI-H322, and NCI-H1404 cell lines; and both cell types with Clara cell granules predominating in the NCI-H441-4 and NCI-H1334 cell lines (9). The differentiation characteristics of Clara cell type and type II alveolar epithelial cell type also have been observed in NCI-H358 and A549 cells, respectively (6-11). Under electron microscopy, we confirmed that all six cell lines conformed to the characteristics of lung adenocarcinoma. The features of Clara cell differentiation were observed only in NCI-H358 cells, whereas differential characteristics of type II alveolar epithelial cell type were observed in A549, NCI-H1975, and HCC827 cells. No differential characteristics of the six cytological subtypes were observed in NCI-H1395 and NCI-H1650 cells, which might be caused by loss of some of the original cell structures during continuous passage in culture and sample processing for electron microscopy.
By comparing DEPs that are expressed significantly different between NCI-H358 cells (Clara cell type) and A549 cells (type II alveolar epithelial cell type) according to 2D-DIGE and MALDI-TOF-MS/MS experiments, a total of five proteins (UCHL1, CK19, CK8, ERO1L, and PRDX2) were identified. Among them, UCHL1 was not verified by western blot, and CK19, CK8, and PRDX2 were clearly differentially expressed between the two cell lines; however, there was no strong-or-nothing expression pattern. Previous studies also have revealed high expression levels of CK19 and CK8 in almost all non-small cell lung cancer cells (16-20), so CK19 and CK8 appeared to be suitable as biomarkers for non-small cell lung cancer cells but not for Clara cell-type lung cancer cells and were excluded from further study. ERO1L was uniquely expressed at a high level in NCI-H358 cells but not in A549 cells. Therefore, the expression level of ERO1L was further validated in six lung cancer cell lines (NCI-H1650, HCC827, NCI-H1395, NCI-H1975, A549, and NCI-H358) by western blot. The ultrastructures of these lung cancer cell lines with high expression of ERO1L were also investigated under electron microscopy. The results showed that ERO1L was nonspecifically overexpressed in cancer cells of Clara cell type and underexpressed in cancer cells of type II alveolar epithelial cell type. Therefore, the present study failed to identify a biomarker by 2D-DIGE and MALDI-TOF-MS/MS. This failure might partially be caused by 2D-DIGE and the target protein. 2D-DIGE is highly sensitive to protein separation of microgram and even nanogram amounts of protein, but it is unable to detect proteins with an extreme isoelectric point, too large or too small of a molecular weight, or a low abundance as well as membrane proteins (21-23). In addition, the fluorescent labeling efficiency is affected by the lysine content of the target protein because the fluorescent dye labels lysine residues of the protein (24,25). Increasing attention has been paid to reverse-phase protein array that is one of antibody-based proteomics modalities for cancer biomarker discovery with higher sensitivity compared with 2D-DIGE and mass spectrometry (26-28).
In the present study, CC10 and SP-C were not DEPs found by 2D-DIGE, but previous studies have suggested the possibility of CC10 and SP-C as biomarkers for distinguishing Clara cell and type II alveolar epithelial cell-type tumors (29,30); therefore, we tested these proteins in the six lung cancer cell lines. Several studies have reported that the existence of CC10 in Clara granules of normal Clara cells, which are speculated as a progenitor of Clara cell-type cancer cells (12-14,31). Other studies have reported infrequent expression of CC10 in human non-small cell lung cancers (29) and positive expression in only 10.2% of lung adenocarcinomas (30), which was less than expected based on previous ultrastructural reports (32). Thus, it is still questionable whether CC10 is suitable as a biomarker for Clara cell-type lung adenocarcinoma cells. SP-A, SP-B, and SP-D are synthesized by type II alveolar epithelial cells and Clara cells, while SP-C is produced only by type II alveolar epithelial cells in normal lungs (10,33,34). To the best of our knowledge, SP-C as a theoretical biomarker for the identification of type II alveolar epithelial cancer cells has not been verified yet. Our western blot and electron microscopy results confirmed that both CC10 and SP-C could not be used as promising biomarkers for classification of Clara cell and type II alveolar epithelial cell-type cancer cells. CC10 was nonspecifically overexpressed in Clara cell-type cancer cells, whereas SP-C was nonspecifically overexpressed in type II alveolar epithelial cell-type cancer cells.
ERO1L is an essential, endoplasmic reticulum-resident protein that plays a widespread role in disulfide-linked protein folding and regulation of a redox state of various proteins in the endoplasmic reticulum (35,36). Patients with early-stage lung adenocarcinoma who had overexpression of ERO1L were positively correlated with a poor survival; and knockdown of ERO1L reduced the viability and migration ability of lung adenocarcinoma cells (37). ERO1L was significantly upregulated in pancreatic cancer cells and patients with high ERO1L expression had short survival time (38). Therefore, ERO1L is associated with poor prognosis of some tumors. In the present study, ERO1L was strongly expressed in NCI-H1975, NCI-H358, and HCC827 cells but weakly expressed in NCI-H1650 cells. Interestingly, all the cells with a strong expression of ERO1L also expressed programmed death-ligand 1 (39,40). The expression characteristics of the strong-or-nothing pattern of ERO1L in lung adenocarcinoma cells might be caused by its suddenly lost expression after some biological processes, suggesting that ERO1L is not suitable as a biomarker for Clara cell-type lung cancer cells but that it can possibly indicate certain biological behaviors of lung cancer cells.
Conclusions
In conclusion, we found that NCI-H1975 and HCC827 cells had characteristics of type II alveolar epithelial cells. We identified a total of five DEPs using 2D-DIGE and MALDI-TOF-MS/MS in the present study, but none of them showed promise as a biomarker to distinguish Clara cell and type II alveolar epithelial cell-type lung adenocarcinoma cells as determined by western blot and electron microscopy.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study does not need ethics approval because of no animals or people involved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Reyes CV, Jensen JD, Graham G. Adenocarcinoma of the lung: electron microscopy of fine-needle aspiration biopsy specimens--a review of 73 cases. Diagn Cytopathol 1999;20:257-60. [Crossref] [PubMed]
- Noguchi M, Shimosato Y. Pulmonary Neoplasms. In: Carter D, Greenson JK, Reuter VE, et al. editors. Sternberg's Diagnostic Surgical Pathology, 5th Edition. Philadelphia: Lippincott Williams & Wilkins, 2009:1066-70.
- Bombi JA, Martinez A, Ramirez J, et al. Ultrastructural and molecular heterogeneity in non-small cell lung carcinomas: study of 110 cases and review of the literature. Ultrastruct Pathol 2002;26:211-8. [Crossref] [PubMed]
- Li G, Chang M, Jiang H, et al. Proteomics analysis of methylglyoxal-induced neurotoxic effects in SH-SY5Y cells. Cell Biochem Funct 2011;29:30-5. [Crossref] [PubMed]
- You J, Wang J, Xie L, et al. D-4F, an apolipoprotein A-I mimetic, inhibits TGF-beta1 induced epithelial-mesenchymal transition in human alveolar epithelial cell. Exp Toxicol Pathol 2016;68:533-41. [Crossref] [PubMed]
- Liu PL, Chong IW, Lee YC, et al. Anti-inflammatory Effects of Resveratrol on Hypoxia/Reoxygenation-Induced Alveolar Epithelial Cell Dysfunction. J Agric Food Chem 2015;63:9480-7. [Crossref] [PubMed]
- Ryndak MB, Singh KK, Peng Z, et al. Transcriptional Profiling of Mycobacterium tuberculosis Replicating in the Human Type II Alveolar Epithelial Cell Line, A549. Genom Data 2015;5:112-4. [Crossref] [PubMed]
- Gazdar AF, Linnoila RI, Kurita Y, et al. Peripheral airway cell differentiation in human lung cancer cell lines. Cancer Res 1990;50:5481-7. [PubMed]
- Canella R, Martini M, Cavicchio C, et al. Involvement of the TREK-1 channel in human alveolar cell membrane potential and its regulation by inhibitors of the chloride current. J Cell Physiol 2019;234:17704-13. [Crossref] [PubMed]
- Kato A, Okura T, Hamada C, et al. Cell stress induces upregulation of osteopontin via the ERK pathway in type II alveolar epithelial cells. PLoS One 2014;9:e100106. [Crossref] [PubMed]
- Wong AP, Keating A, Waddell TK. Airway regeneration: the role of the Clara cell secretory protein and the cells that express it. Cytotherapy 2009;11:676-87. [Crossref] [PubMed]
- Méndez A, Rojas DA, Ponce CA, et al. Primary infection by Pneumocystis induces Notch-independent Clara cell mucin production in rat distal airways. PLoS One 2019;14:e0217684. [Crossref] [PubMed]
- An JY, Ahn C, Kang HY, et al. Inhibition of mucin secretion via glucocorticoid-induced regulation of calcium-related proteins in mouse lung. Am J Physiol Lung Cell Mol Physiol 2018;314:L956-66. [Crossref] [PubMed]
- Nureki SI, Tomer Y, Venosa A, et al. Expression of mutant Sftpc in murine alveolar epithelia drives spontaneous lung fibrosis. J Clin Invest 2018;128:4008-24. [Crossref] [PubMed]
- Ueda Y, Fujita J, Bandoh S, et al. Expression of cytokeratin 19 mRNA in human lung cancer cell lines. Int J Cancer 1999;81:939-43. [Crossref] [PubMed]
- Masai K, Nakagawa K, Yoshida A, et al. Cytokeratin 19 expression in primary thoracic tumors and lymph node metastases. Lung Cancer 2014;86:318-23. [Crossref] [PubMed]
- Yan L, Yao Y, Wang LH, et al. Detection of CK19, LUNX, and KS1/4 mRNA expression in the peripheral blood for diagnosis of micrometastases in patients with non-small cell lung cancer and their clinical implications. Genet Mol Res 2015;14:15090-5. [Crossref] [PubMed]
- Gharib TG, Chen G, Wang H, et al. Proteomic analysis of cytokeratin isoforms uncovers association with survival in lung adenocarcinoma. Neoplasia 2002;4:440-8. [Crossref] [PubMed]
- Serpil O, Meral A, Muzeyyen O, et al. Alkaline phosphatase, cytokeratin 7, cytokeratin 8 in the diagnosis of primary lung adenocarcinoma from 148 pleura fluids specimens. Folia Histochem Cytobiol 2009;47:87-92. [Crossref] [PubMed]
- Bae SH, Harris AG, Hains PG, et al. Strategies for the enrichment and identification of basic proteins in proteome projects. Proteomics 2003;3:569-79. [Crossref] [PubMed]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis 2000;21:1054-70. [Crossref] [PubMed]
- Gygi SP, Corthals GL, Zhang Y, et al. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A 2000;97:9390-5. [Crossref] [PubMed]
- Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 1997;18:2071-7. [Crossref] [PubMed]
- Arentz G, Weiland F, Oehler MK, et al. State of the art of 2D DIGE. Proteomics Clin Appl 2015;9:277-88. [Crossref] [PubMed]
- Kondo T. Cancer biomarker development and two-dimensional difference gel electrophoresis (2D-DIGE). Biochim Biophys Acta Proteins Proteom 2019;1867:2-8.
- Peng J, Zhang J, Zou D, et al. Proteomics score: a potential biomarker for the prediction of prognosis in non-small cell lung cancer. Transl Cancer Res 2019;8:1904-17. [Crossref]
- Rader JS, Pan A, Corbin B, et al. Identification and validation of a prognostic proteomic signature for cervical cancer. Gynecol Oncol 2019;155:324-30. [Crossref] [PubMed]
- Linnoila RI, Szabo E, DeMayo F, et al. The role of CC10 in pulmonary carcinogenesis: from a marker to tumor suppression. Ann N Y Acad Sci 2000;923:249-67. [Crossref] [PubMed]
- Nomori H, Morinaga S, Kobayashi R, et al. Protein 1 and Clara cell 10-kDa protein distribution in normal and neoplastic tissues with emphasis on the respiratory system. Virchows Arch 1994;424:517-23. [Crossref] [PubMed]
- Leeman KT, Fillmore CM, Kim CF. Lung stem and progenitor cells in tissue homeostasis and disease. Curr Top Dev Biol 2014;107:207-33. [Crossref] [PubMed]
- Broers JL, Jensen SM, Travis WD, et al. Expression of surfactant associated protein-A and Clara cell 10 kilodalton mRNA in neoplastic and non-neoplastic human lung tissue as detected by in situ hybridization. Lab Invest 1992;66:337-46. [PubMed]
- Kuroki Y, McCormack FX, Ogasawara Y, et al. Epitope mapping for monoclonal antibodies identifies functional domains of pulmonary surfactant protein A that interact with lipids. J Biol Chem 1994;269:29793-800. [PubMed]
- Khan P, Fytianos K, Tamo L, et al. Culture of human alveolar epithelial type II cells by sprouting. Respir Res 2018;19:204. [Crossref] [PubMed]
- Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1998;1:171-82. [Crossref] [PubMed]
- Kutomi G, Tamura Y, Tanaka T, et al. Human endoplasmic reticulum oxidoreductin 1-alpha is a novel predictor for poor prognosis of breast cancer. Cancer Sci 2013;104:1091-6. [Crossref] [PubMed]
- Hsu CH, Hsu CW, Hsueh C, et al. Identification and Characterization of Potential Biomarkers by Quantitative Tissue Proteomics of Primary Lung Adenocarcinoma. Mol Cell Proteomics 2016;15:2396-410. [Crossref] [PubMed]
- Han F, Xu Q, Zhao J, et al. ERO1L promotes pancreatic cancer cell progression through activating the Wnt/catenin pathway. J Cell Biochem 2018;119:8996-9005. [Crossref] [PubMed]
- Jiang XM, Xu YL, Huang MY, et al. Osimertinib (AZD9291) decreases programmed death ligand-1 in EGFR-mutated non-small cell lung cancer cells. Acta Pharmacol Sin 2017;38:1512-20. [Crossref] [PubMed]
- Suda K, Rozeboom L, Rivard CJ, et al. Therapy-induced E-cadherin downregulation alters expression of programmed death ligand-1 in lung cancer cells. Lung Cancer 2017;109:1-8. [Crossref] [PubMed]


