
This article has an erratum available at: http://dx.doi.org/10.21037/tcr-2020-008
The prognosis analysis of RFWD2 inhibiting the expression of ETV1 in colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the common gastrointestinal malignancies, its incidence ranks third in the world, the tumor-related lethality ranks fourth in the world. CRC is a heterogeneous disease that caused by the interaction of genetic and environmental factors (1,2). CRC is one of the most prevalent and deadly incident cancers worldwide (3,4). In recent years, with the improvement of living standards, the incidence and mortality of CRC in China gradually is increasing (5). With improving diagnostic techniques, and by surgery and chemotherapy for CRC had better effect on early treatment (6). However, for advanced CRC patients, due to the chemotherapy-insensitive and easy to relapse lead to the five year survival rate is only 50% (7). Tumor metastasis and recurrence is the most important factor affecting the survival rate of CRC patients. Therefore, studying the molecular mechanism of CRC metastasis and looking for new targets for prevention and treatment are still the hot topics in the field of cancer research.
RFWD2 also known as COP1 (constitutive photomorphogenic 1) protein, comprising RING finger, coiled-coil and WD40 domains, is conserved in both higher plants and vertebrates (8). Scientists have pointed at possible roles for mammalian COP1 in tumorigenesis and the stress response through regulating the activities of p53 (9) and c-Jun (10). Recent genetic studies have shown that RFWD2 deficiency leads to spontaneous tumor formation in mice, and have identified mutations in RFWD2 and its substrates in various human cancers (11). RFWD2 was identified with relevant roles in tumorigenesis process.
The oncoproteins ETV family is implicated in melanomas, breast and other types of cancer. Complex post-translational modifications govern the activity of PEA3 factors, which promote cell proliferation, motility and invasion (12). The ETS family is one of the largest families of signal-dependent transcriptional regulators (13). It is involved in tumorigenesis and development of a variety of tumors (14). The ETS family is overexpressed in breast cancer (15), prostate cancer (16), melanoma (17), Ewing’s tumor (18) and gastrointestinal stromal tumor (19) and also involved in the development of tumors.
Herein, we determined the expression of RFWD2 and ETV1 in CRC through bioinformatic analysis, aiming to ascertain whether they are potential molecular biomarkers for the early diagnosis of CRC and to obtain clues for the pathogenesis of CRC.
Methods
Ethics statement
The research was approved by the Ethics Committee of Guangzhou Medical University (No. 2018-KY-106C), and written informed consent was obtained from each patient involved in the study.
Immunohistochemical analysis
Archived and paraffin-embedded samples were obtained from 76 CRC patients who underwent surgical resection between January 2013 and March 2019 in the Second Affiliated Hospital of Guangzhou University and the Affiliated Shunde Hospital of Guangzhou Medical University. Immunohistochemical staining of ETV1 or RFWD2 was carried out according to the manufacture’s protocol. In briefly, the sections were incubated overnight in a moist box with antibodies of ETV1 (1:100; ABCAM, USA) or RFWD2 (1:200; ABCAM, USA) in PBS at 4 °C. Poly peroxidase anti-mouse/rabbit IgG (GSGB-BIO, China) was subjected to the sections for 30 minutes at room temperature after washing with PBS. Diaminobenzidine was used for colorimetric detection and the sections were counterstained with haematoxylin and mounted with distyrene plasticizer xylene (DPX). Negative controls were performed by replacing the primary antibody with preimmune rabbit serum. Positive controls were conducted according to the manufacture’s suggestion. For each run of immunohistochemistry, negative and positive controls were performed. Immunostained tissue array sections were reviewed under a microscope by two pathologists, who were blinded regarding the clinicopathological characteristics and outcome of the patients, while visually scoring each individual tissue core. For positively stained cells: 0 (negative) was denoted for <10% positive cells, 1 (weak) for <25% positive cells, 2 (moderate) for <50% positive cells, and 3 (strong) for >50% positive cells. The staining intensity was defined as: 0 for no stain, 1 for weak-positive (faint yellow), 2 for moderate-positive (yellowish-brown), and 3 for strong-positive (brown). Scores of the proportion and intensity of positively stained tumor cells were added and stratified as having negative (−) expression (0–3 score) and positive (+) expression (4–6 score).
Bioinformatic analysis of RFWD2 and ETV1
The human patients’ samples from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) were analyzed to investigate the clinical significance of RFWD2 and ETV1 expression in CRC patients. Cancer Cell Line Encyclopedia databases (CCLE, https://portals.broadinstitute.org/ccle) was used to investigate the expression levels of RFWD2 and ETV1. In Linkedomics (http://www.linkedomics.org/) we dug out the positively and negatively associated genes regulated by RFWD2 and ETV1. To explore the functional annotation and pathway enrichment of RFWD2 and ETV1 genes, the Gene Ontology (GO; http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/kegg/kegg2.html) database analyses were conducted using a Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 online analysis tool with P<0.05 as the significant threshold to obtain significant gene sets.
Statistical analysis
Data were expressed as the mean ± SD from at least three independent experiments for each group. The differences between RFWD2 or ETV1 expression levels with the clinicopathological features groups were analyzed using a factorial model one-way analysis of variance. All statistical analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, IL, USA) and P less than 0.05 was considered statistically significant.
Results
Association of RFWD2 and ETV1 expression with clinicopathological characteristics in CRC patients
The human patients’ samples from TCGA were analyzed to investigate the clinical significance of RFWD2 and ETV1 expression levels in CRC patients. In Figure 1, the results showed that RFWD2 expression was obviously different in major cancer stage (P<0.001; Figure 1A) and individual cancer stages (P=0.0057 normal vs. stage 1; P<0.001, normal vs. stage 2; P=0.0132 normal vs. stage 3; Figure 1B). Both gender showed the significance in RFWD2 expression compared with normal (P=0.0066 normal vs. male; P<0.001, normal vs. female; Figure 1C). Interestingly, the patients’ weight whether normal or overweight were all associated with RFWD2 protein expression (P=0.0102 normal vs. normal weight; P=0.0064 normal vs. extreme weight; P=0.0396 normal vs. obese; P=0.0028 normal vs. extreme obese; Figure 1D). The other characteristics, such as age and race were also associated with RFWD2 protein expression (P=0.0010, normal vs. Caucasian; P<0.001, normal vs. African American; Figure 1E. P<0.001, normal vs. 41–60 years; P<0.001, normal vs. 61–80 years; Figure 1F).
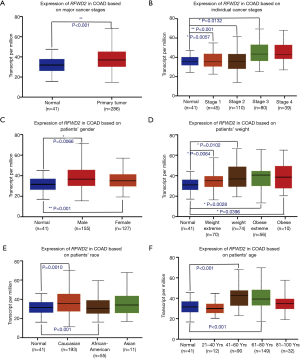
As summarized in Figure 2, the expression level of ETV1 had significant differences in major cancer stage (P=0.0349; Figure 2A) and individual cancer stages (P=0.0072 normal vs. stage 3; P=0.0229 stage 1 vs. stage 3; Figure 2B), female patients had more ETV1 expression than normal (P=0.0313; Figure 2C). The ETV1 expression levels of extreme obese patients and Caucasian people were higher compared with normal respectively (P=0.0152; Figure 2D; P=0.0166; Figure 2E), but not in the patients’ age (Figure 2F).
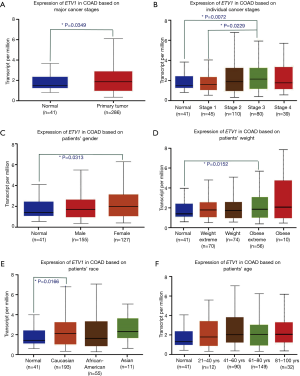
Expression and correlation analysis of RFWD2 and ETV1 in CRC tissues and cell lines
In addition, the results of Cancer Cell Line Encyclopedia databases was consistent with that of TCGA, demonstrating the mRNA expression level of RFWD2 was more than 1.8 times of ETV1 (Figure 3A,B), the spearman correlative analysis showed that their expression was negative relevant in colorectal tumor tissue (r2=0.2015, F=14.13, P<0.001, Figure 3C).
Immunohistochemical and prognosis Analysis of RFWD2 and ETV1 in CRC patients
Among the 76 cases of CRC 25 cases (32.9%) were RFWD2-positive and 51 (67.1%) were RFWD2-negative; whereas 47 cases (61.8%) were ETV1-positive and 29 (38.2%) were ETV1-negative. Representative images of immunohistochemical analysis of RFWD2 and ETV1 in CRC patients were showed in Figure 4. ETV1 expression status according to RFWD2 expression and their corresponding clinicopathological characteristics were summarized in Table 1. Either ETV1 or RFWD2 expression status was significantly associated with TNM stages of CRC patients, the number of lymph nodes involved and tumor relapse.
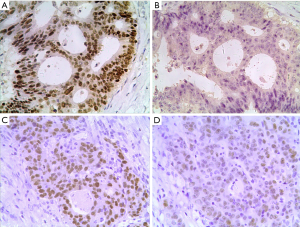
Table 1
| Characteristics | RFWD2 | P value | ETV1 | P value | ||
|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | |||
| Tumor size (cm) | ||||||
| ≤3 | 11 | 4 | 0.3346 | 4 | 13 | 0.3821 |
| >3 | 50 | 11 | 10 | 49 | ||
| Pathological type | ||||||
| COAD | 44 | 23 | 0.3769 | 27 | 39 | 0.1816 |
| Other | 7 | 2 | 2 | 8 | ||
| TNM stage | ||||||
| I | 4 | 2 | 0.0007 | 1 | 2 | 0.0093 |
| II | 10 | 6 | 10 | 19 | ||
| III | 52 | 2 | 3 | 41 | ||
| Lymph node metastasis | ||||||
| 0–4 | 37 | 5 | 0.0004 | 10 | 23 | 0.0478 |
| 5–8 | 12 | 14 | 2 | 29 | ||
| ≥9 | 7 | 1 | 2 | 10 | ||
| Relapse | ||||||
| No | 2 | 50 | 0.0000 | 11 | 15 | 0.0003 |
| Yes | 15 | 9 | 3 | 47 | ||
RFWD2, E3 ubiquitin ligase Ring finger and WD domain 2; ETV1, ETS variant 1.
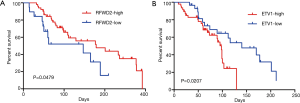
Then, the proteins expression status of RFWD2 or ETV1 in CRC tissues was investigated for associations with overall survival by using logrank test for significance estimates. As indicated by the results, patients with high RFWD2 expression showed a significantly longer cancer-specific survival than those with a low level of RFWD2 expression (P=0.0479; Figure 5A). However, patients with high ETV1 expression had a markedly poor overall survival compared to patients with low ETV1 expression (P=0.0207; Figure 5B). These results suggested that RFWD2 and ETV1 might have the opposite relationship in tumorigenesis process.
The gene enrichment analysis of RFWD2 and ETV1 in CRC patients
Since RFWD2 and ETV1 had the opposite relationship in expression and function as mentioned before, we next performed further exploration on the potential roles of ETV1 and RFWD2 in CRC, in connection with other featured biomarkers. In Linkedomics website, the positively and negatively associated genes regulated by RFWD2 and ETV1 were dug out.
To systematically summarize RFWD2 and ETV1 gene function, Gene ontology enrichment analysis was performed to analyze RFWD2 and ETV1 associated genes. Nine cellular component GO terms of RFWD2 biological process focused on: interleukin-17 production; peptide cross-linking; amino sugar metabolic process; ATP hydrolysis coupled transmembrane transport; type 2 immune response; dopamine receptor signaling pathway; post-translational protein modification and protein localization to Golgi apparatus; oligosaccharide-lipid intermediate biosynthetic process (Table 2). And the biological process of ETV1 may involve in nucleotide phosphorylation; secondary metabolic process; columnar/cuboidal epithelial cell differentiation; platelet-derived growth factor receptor signaling pathway; mesenchymal cell proliferation; spinal cord development; odontogenesis; regulation of behavior; central nervous system neuron differentiation; interleukin-13 production and response to osmotic stress (Table 3).
Table 2
| Gene set | Description | Size | P value |
|---|---|---|---|
| GO:0032620 | Interleukin-17 production | 25 | 0e+00 |
| GO: 0018149 | Peptide cross-linking | 56 | 0e+00 |
| GO:0006040 | Amino sugar metabolic process | 38 | 4.06e-03 |
| GO:0090662 | ATP hydrolysis coupled transmembrane transport | 34 | 2.04e-03 |
| GO:0042092 | Type 2 immune response | 33 | 3.06e-02 |
| GO:0007212 | Dopamine receptor signaling pathway | 37 | 3.25e-02 |
| GO:0043687 | Post-translational protein modification | 39 | 0e+00 |
| GO:0034067 | Protein localization to Golgi apparatus | 34 | 0e+00 |
| GO:0006490 | Oligosaccharide-lipid intermediate biosynthetic process | 21 | 4e-02 |
RFWD2, E3 ubiquitin ligase Ring finger and WD domain 2.
Table 3
| Gene set | Description | Size | P value |
|---|---|---|---|
| GO:0046939 | Nucleotide phosphorylation | 88 | 8e-03 |
| GO:0019748 | Secondary metabolic process | 55 | 1.6e-02 |
| GO:0002065 | Columnar/cuboidal epithelial cell differentiation | 97 | 2e-03 |
| GO:0060021 | Palate development | 85 | 8e-03 |
| GO:0048008 | Platelet-derived growth factor receptor signaling pathway | 46 | 3.41e-02 |
| GO:0010463 | Mesenchymal cell proliferation | 47 | 2.4e-02 |
| GO:0021510 | Spinal cord development | 93 | 8e-03 |
| GO:0042476 | Odontogenesis | 113 | 0e+00 |
| GO:0050795 | Regulation of behavior | 60 | 2.4e-02 |
| GO:0021953 | Central nervous system neuron differentiation | 168 | 0e+00 |
| GO:0032616 | Interleukin-13 production | 20 | 0e+00 |
| GO:0006970 | Response to osmotic stress | 65 | 3.2e-02 |
ETV1, ETS variant 1.
Analysis of cellular components enrichment analysis suggested that RFWD2 may be expressed in telomerase holoenzyme complex; ATPase complex; cornified envelope; PcG protein complex and membrane region (Table 4). ETV1 may be expressed in GTPase complex; sperm part; replication fork; cell projection membrane; cytoplasmic region; secretory vesicle; intrinsic component of organelle membrane; cytoplasmic vesicle membrane; endoplasmic reticulum lumen and cell surface furrow (Table 5).
Table 4
| Gene set | Description | Size | P value |
|---|---|---|---|
| GO:0005697 | Telomerase holoenzyme complex | 21 | 4.19e-02 |
| GO:1904949 | ATPase complex | 23 | 1.03e-02 |
| GO:0001533 | Cornified envelope | 46 | 0e+00 |
| GO:0031519 | PcG protein complex | 23 | 2.42e-02 |
| GO:0098589 | Membrane region | 347 | 1.8e-02 |
RFWD2, E3 ubiquitin ligase Ring finger and WD domain 2.
Table 5
| Gene set | Description | Size | P value |
|---|---|---|---|
| GO:1905360 | GTPase complex | 31 | 6e-03 |
| GO:0097223 | Sperm part | 151 | 0e+00 |
| GO:0005657 | Replication fork | 63 | 4.2e-02 |
| GO:0031253 | Cell projection membrane | 289 | 2.8e-02 |
| GO:0099568 | Cytoplasmic region | 286 | 4.2e-02 |
| GO:0099503 | Secretory vesicle | 454 | 6e-03 |
| GO:0031300 | Intrinsic component of organelle membrane | 154 | 4.2e-02 |
| GO:0030659 | Cytoplasmic vesicle membrane | 465 | 2e-03 |
| GO:0005788 | Endoplasmic reticulum lumen | 195 | 2.4e-02 |
| GO:0097610 | Cell surface furrow | 46 | 3.8e-02 |
ETV1, ETS variant 1.
Analysis of Molecular Function demonstrated that RFWD2 may participate in ephrin receptor binding; sulfur compound transmembrane transporter activity; extracellular matrix structural constituent and virus receptor activity (Table 6), and ETV1 may participate in taste receptor activity; Wnt-activated receptor activity; GTP-dependent protein binding; macromolecule transmembrane transporter activity; carbohydrate kinase activity; semaphorin receptor binding; drug transporter; E-box binding; actinin binding; cyclin binding; phosphotransferase activity, phosphate group as acceptor; cyclin-dependent protein kinase activity and oxidoreductase activity, acting on single donors with incorporation of molecular oxygen (Table 7).
Table 6
| Gene set | Description | Size | P value |
|---|---|---|---|
| GO:0046875 | Ephrin receptor binding | 26 | 2.06e-03 |
| GO:1901682 | Sulfur compound transmembrane transporter activity | 27 | 4.53e-02 |
| GO:0005201 | Extracellular matrix structural constituent | 75 | 1e-02 |
| GO:0001618 | Virus receptor activity | 65 | 1.4e-02 |
RFWD2, E3 ubiquitin ligase Ring finger and WD domain 2.
Table 7
| Gene set | Description | Size | P value |
|---|---|---|---|
| GO:0008527 | Taste receptor activity | 29 | 0e+00 |
| GO:0042813 | Wnt-activated receptor activity | 22 | 2.21e-02 |
| GO:0030742 | GTP-dependent protein binding | 24 | 1.42e-02 |
| GO:0022884 | Macromolecule transmembrane transporter activity | 21 | 2.43e-02 |
| GO:0019200 | Carbohydrate kinase activity | 20 | 3.23e-02 |
| GO:0030215 | Semaphorin receptor binding | 23 | 4.86e-02 |
| GO:0090484 | Drug transporter | 21 | 3.64e-02 |
| GO:0070888 | E-box binding | 33 | 6.04e-03 |
| GO:0042805 | Actinin binding | 29 | 4.61e-02 |
| GO:0030332 | Cyclin binding | 20 | 3.22e-02 |
| GO:0016776 | Phosphotransferase activity, phosphate group as acceptor | 39 | 2.2e-02 |
| GO:0097472 | Cyclin-dependent protein kinase activity | 32 | 4.01e-02 |
| GO:0016701 | Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | 26 | 0e+00 |
ETV1, ETS variant 1.
KEGG datasets revealed that RFWD2 might involve in Asthma; Fat digestion and absorption and Starch and sucrose metabolism (Table 8), and ETV1 might involve in Neomycin, kanamycin and gentamicin biosynthesis; Sulfur metabolism; Mismatch repair; Selenocompound metabolism; Mannose type O-glycan biosynthesis; Galactose metabolism; Tryptophan metabolism; Glycine, serine and threonine metabolism and Viral myocarditis (Table 9). Therefore, our next step research will focus on the above bioinformatics data to investigate the function and mechanism of RFWD2 and ETV1 in CRC cells.
Table 8
| Gene set | Description | Size | P value | Leading edge gene |
|---|---|---|---|---|
| hsa05310 | Asthma | 16 | 1.98e-02 | FCER1A; FCER1G; IL10 |
| hsa04975 | Fat digestion and absorption | 38 | 0e+00 | PLA2G2D; PLA2G2E; APOA1; APOA4; PLA2G2C; PLA2G2A; PLA2G5; PLA2G2F; MOGAT2; DGAT2 |
| hsa00500 | Starch and sucrose metabolism | 35 | 0e+00 | TREH; AGL; GBE1; AMY1A; AMY1B; AMY1C; AMY2A; AMY2B; PGM2L1; PGM1; PYGM |
RFWD2, E3 ubiquitin ligase Ring finger and WD domain 2.
Table 9
| Gene set | Description | Size | P value | Leading edge gene |
|---|---|---|---|---|
| hsa00524 | Neomycin, kanamycin and gentamicin biosynthesis | 5 | 2.42e-03 | GCK |
| hsa00920 | Sulfur metabolism | 10 | 2.36e-02 | CYCS |
| hsa03430 | Mismatch repair | 23 | 6.05e-03 | PMS2; POLD2; RFC2; RPA3; SSBP1 |
| hsa00450 | Selenocompound metabolism | 15 | 1.65e-02 | INMT |
| hsa00515 | Mannose type O-glycan biosynthesis | 18 | 2.42e-02 | ISPD |
| hsa00052 | Galactose metabolism | 30 | 2.2e-02 | AKR1B1; GCK; AKR1B10; MGAM |
| hsa00380 | Tryptophan metabolism | 38 | 4.01e-03 | INMT; DDC; AOC1; OGDH |
| hsa00260 | Glycine, serine and threonine metabolism | 39 | 0e+00 | DLD; PGAM2; PSPH; BPGM |
| hsa05416 | Viral myocarditis | 38 | 1e-02 | CYCS; RAC1; ACTB; CAV1; CD40 |
ETV1, ETS variant 1.
Discussion
Colon and rectum cancer (CRC) is the second most lethal type of cancer in United States (20). Therefore, identification of the CRC-specific biomarkers involved in these procedures is very important for diagnosis, therapy and prognostic prediction in clinics.
RFWD2 overexpression had been found in many tumor types. High expression of RFWD2 was found in promoting cell proliferation, cell transformation, and tumor progression, manifesting its role as cancer promoter (21). These results suggested that RFWD2 might play an important role in promoting tumorigenesis or progression. These results supported an earlier hypothesis that RFWD2 might be an oncogene. However, recent study showed that RFWD2 might be a tumor suppressor in patients with Triple-negative breast cancer (15). Evidence also showed that loss of COP1 (RFWD2) expression determines poor prognosis in patients with gastric cancer (22). COP1 was downregulated in renal cell carcinoma (RCC) and inhibited the migration of RCC ACHN cells in vitro (23). All above data suggested that RFWD2 was a potent tumor-suppressor or oncogene in different kinds of human cancers, and the expression and clinical significance of RFWD2 in CRC had not been explored.
ETV1 was found significantly associated with lymphatic metastasis of CRC (22). miR-17-5p acts as a tumor suppressor in Triple-negative breast cancer by targeting ETV1, and a low-abundance of miR-17-5p may be involved in the pathogenesis of Triple-negative breast cancer (24). ETV1 and other Pea3-ETS transcription factors are critical nuclear effectors of MAPK signaling that are regulated through protein stability. COP1-DET1-ETS axis played an important role in regulating ERK transcriptome and sensitivity to MAPK inhibitors (25).
In our study, CRC patients with high RFWD2 expression showed a significantly longer cancer-specific survival than those with a low level of RFWD2 expression. While patients with high ETV1 expression had a markedly poor overall survival compared to patients with low ETV1 expression. These results indicated that RFWD2 and ETV1 might have the opposite relationship in function. Results from our collected data showed that among the 76 cases of CRC 25 cases (32.9%) were RFWD2-positive and 51 (67.1%) were negative; whereas 47 cases (61.8%) were ETV1-positive and 29 (38.2%) were ETV1-negative. The spearman correlative analysis showed that their expression was negative relevant in colorectal tumor tissue. Either ETV1 or RFWD2 expression status was significantly associated with TNM stages of CRC patients, the number of lymph nodes involved and tumor relapse.
Our study provides a new insight on functional roles of ETV1 or RFWD2 gene in CRC. According to the predicting results obtained from GO bioinformatics, we detected that RFWD2 could involve in interleukin-17 production; peptide cross-linking; amino sugar metabolic process; ATP hydrolysis coupled transmembrane transport; type 2 immune response; dopamine receptor signaling pathway; post-translational protein modification and protein localization to Golgi apparatus; oligosaccharide-lipid intermediate biosynthetic process, suggested that RFWD2 could participate in ephrin receptor binding; sulfur compound transmembrane transporter activity; extracellular matrix structural constituent and virus receptor activity. In addition, Kyoto Encyclopedia of Genes and Genomes datasets (KEGG) suggested that RFWD2 could target on Asthma; Fat digestion and absorption and Starch and sucrose metabolism. Meanwhile ETV1 could target on Neomycin, kanamycin and gentamicin biosynthesis; Sulfur metabolism; Mismatch repair; Selenocompound metabolism; Mannose type O-glycan biosynthesis; Galactose metabolism; Tryptophan metabolism; Glycine, serine and threonine metabolism and Viral myocarditis. These pathways predicted from RFWD2 and ETV1 were related to metabolism, survival, proliferation and transcription regulation, respectively, which might involve in the potential regulating function in CRC.
Conclusions
In conclusion, we demonstrated that RFWD2 functioning as an oncogene might be a tumor suppressor in negatively regulating ETV1 in patients with CRC. By our data suggested that RFWD2 and ETV1 could have the potential to serve as molecular biomarkers for the development and progression of CRC. It provides important message about effective therapeutic targets towards changing CRC patients' outcome and is worth to be further explored.
Acknowledgments
Funding: This work was funded by
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.35). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This submission has been received explicitly from all co-authors. And authors whose names appear on the submission have contributed sufficiently to the scientific work and therefore share collective responsibility and accountability for the results. Approval for this study was obtained by Ethics Committee of Guangzhou Medical University and the ethics committee waived the use of the inform consent (these CRC tumor specimens are the remaining tissues after pathological examination. This study does not cause any secondary damage to the patients.).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Subramanian S, Bobashev G, Morris RJ, et al. Personalized medicine for prevention: can risk stratified screening decrease colorectal cancer mortality at an acceptable cost? Cancer Causes Control 2017;28:299-308. [Crossref] [PubMed]
- Binefa G, Rodriguez-Moranta F, Teule A, et al. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol 2014;20:6786-808. [Crossref] [PubMed]
- Dickinson BT, Kisiel J, Ahlquist DA, et al. Molecular markers for colorectal cancer screening. Gut 2015;64:1485-94. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Lai YL, Lin JK, Liang WY, et al. Surgical resection combined with chemotherapy can help achieve better outcomes in patients with primary colonic lymphoma. J Surg Oncol 2011;104:265-8. [Crossref] [PubMed]
- Adam R, Haller DG, Poston G, et al. Toward optimized front-line therapeutic strategies in patients with metastatic colorectal cancer--an expert review from the International Congress on Anti-Cancer Treatment (ICACT) 2009. Ann Oncol 2010;21:1579-84. [Crossref] [PubMed]
- Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 2005;15:618-25. [Crossref] [PubMed]
- Satija YK, Bhardwaj A, Das S. A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer. Int J Cancer 2013;133:2759-68. [PubMed]
- Sanchez-Barcelo EJ, Mediavilla MD, Vriend J, et al. Constitutive photomorphogenesis protein 1 (COP1) and COP9 signalosome, evolutionarily conserved photomorphogenic proteins as possible targets of melatonin. J Pineal Res 2016;61:41-51. [Crossref] [PubMed]
- Marine JC. Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer 2012;12:455-64. [Crossref] [PubMed]
- Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012;1826:1-12.
- Jedlicka P, Gutierrez-Hartmann A. Ets transcription factors in intestinal morphogenesis, homeostasis and disease. Histol Histopathol 2008;23:1417-24. [PubMed]
- Birner P, Berghoff AS, Dinhof C, et al. MAP kinase activity supported by BRAF (V600E) mutation rather than gene amplification is associated with ETV1 expression in melanoma brain metastases. Arch Dermatol Res 2014;306:873-84. [Crossref] [PubMed]
- Ouyang M, Wang H, Ma J, et al. COP1, the negative regulator of ETV1, influences prognosis in triple-negative breast cancer. BMC Cancer 2015;15:132. [Crossref] [PubMed]
- Selvaraj N, Budka JA, Ferris MW, et al. Prostate cancer ETS rearrangements switch a cell migration gene expression program from RAS/ERK to PI3K/AKT regulation. Mol Cancer 2014;13:61. [Crossref] [PubMed]
- Jané-Valbuena J, Widlund HR, Perner S, et al. An oncogenic role for ETV1 in melanoma. Cancer Res 2010;70:2075-84. [Crossref] [PubMed]
- Ordóñez JL, Osuna D, Herrero D, et al. Advances in Ewing's sarcoma research: where are we now and what lies ahead? Cancer Res 2009;69:7140-50. [Crossref] [PubMed]
- Jang BG, Lee HE, Kim WH. ETV1 mRNA is specifically expressed in gastrointestinal stromal tumors. Virchows Arch 2015;467:393-403. [Crossref] [PubMed]
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104-17. [Crossref] [PubMed]
- Dornan D, Bheddah S, Newton K, et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res 2004;64:7226-30. [Crossref] [PubMed]
- Sawada G, Ueo H, Matsumura T, et al. Loss of COP1 expression determines poor prognosisin patients with gastric cancer. Oncol Rep 2013;30:1971-5. [Crossref] [PubMed]
- Ta L, Xuan C, Xing N, et al. COP1 is downregulated in renal cell carcinoma (RCC) and inhibits the migration of RCC ACHN cells in vitro. Mol Med Rep 2016;14:1371-8. [Crossref] [PubMed]
- Li J, Lai Y, Ma J, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer 2017;17:745. [Crossref] [PubMed]
- Xie Y, Cao Z, Wong EW, et al. COP1-DET1-ETS axis regulates ERK transcriptome and sensitivity to MAPK inhibitors. J Clin Invest 2018;128:1442-57. [Crossref] [PubMed]


