
Up-regulation of FAT4 enhances the chemosensitivity of colorectal cancer cells treated by 5-FU
Introduction
Colorectal cancer (CRC) has the fourth highest mortality worldwide, moreover, annually, more than 1.2 million people suffer from CRC (1). 5-fluorouracil (5-FU) is a chemotherapeutic drug widely used as a clinical cancer suppressor in chemotherapy for certain types of cancers, including CRC (2). 5-FU is considered to be a highly efficient chemotherapeutic agent, as it could successfully increase the 12-month survival for CRC patients (3). However, the toxicity of 5-FU limits its doses in clinical treatments and drug resistance developed by tumor cells reduce its effectiveness as a cancer suppressor (4). Thus, further investigation about the mechanism underlying the occurrence of CRC and finding mediating genes were urgently required. In the present, some genes have been found to be able to attenuate the process during which 5-FU suppresses CRC, for example, miRNA-522 was reported to enhance 5-FU resistance by moderating SMAD2 signaling pathway in CRC (5). Through inhibiting the expression of ABC transport protein, lidocaine could increase 5-FU cytotoxicity to choriocarcinoma cells (6).
FAT family, which belongs to the cadherin superfamily, consists of a group of calcium-dependent cell adhesion molecules. Studies showed that FAT family was closely connected to the differentiation of cells and tissues (7), moreover, FAT takes part in the Salvador-Warts-Hippo signaling pathway, which is a widely recognized regulator related to the size of Drosophila organ (8). Furthermore, biological properties of cells in epithelial tissues, which are highly associated with the mutation of cell and tumorigenicity, have been reported to be able to moderate the Salvador-Warts-Hippo signaling pathway (9). Qi proposed that FAT4 could act as a tumor suppressor by observing tumor behavior of breast cancer cells when FAT4 was low-expressed or over-expressed (10). Studies were less conducted on the relationship between FAT4 and CRC, but the correlation between FAT4 and other cancers have been widely investigated. Li reported that FAT4 could act as a mediator in the down-regulation of lncRNA RNAZFHX4-AS1, so as to inhibit the migration and invasion of breast cancer (11). Han investigated the pathogenesis of prostate cancer, and pointed out that the knockdown of FAT4 resulted in a more severe condition of prostate cancer (12). Epithelial-mesenchymal transition (EMT) process refers to a physiological process associated with tumor cell migration and invasion that affect greatly tumor metastasis and development (13). In breast cancer, the knockdown of lncRNA PANDAR was found to inhibit EMT process by down-regulating the expression of EMT-related markers, so as to suppress the cell proliferation, invasion of breast cancer cells (14). Moreover, the EMT process is highly associated with the prognosis of lung cancer patients, therefore is seen a predictor of prognosis of lung cancer patients (15). As mentioned above, FAT family, together with some EMT markers such as E-Cadherin, N-Cadherin, all belong to cadherin family, as a result, the possible relationship between them in the process of cancerous progression requires further investigation.
In the present study, we aimed to explore the role of FAT4 in the cell proliferation, invasion and apoptosis and EMT process of CRC cells under the treatment of 5-FU.
Methods
Formalin-fixed paraffin-embedded human samples
Human CRC and paired adjacent tissue samples (formalin-fixed, paraffin-embedded, FFPE) were derived from 30 CRC patients (19 males and 11 females) who received tumor resection in the First Affiliated Hospital of Zhejiang Chinese Medical University between 01/11/2016 to 29/01/2018. All patients and their relatives were informed and signed the consent form for allowing their tissues to be used in the research. The study was approved by the First Affiliated Hospital of Zhejiang Chinese Medical University Ethics Committee (2019).
Cell lines
Human normal colon cell line CCD-18Co (CRL-1459™), human normal colorectal mucosa cell line FHC (CRL-1831), human CRC cell lines LS174T (CL-188), HT29 (HTB-38), HCT116 (CCL-247), HCT8 (CCL-244), HCT-15 (CCL-225), SW-620 (CCL-227) and LOVO (CCL-229) were purchased from American Type Culture Collection (ATCC, USA). All the cells were cultured in a humidified cell incubator with 5% CO2 at 37 °C. FHC cells were cultured in MEM (32561037, Gibco, ThermoFisher, USA) containing 10% Fetal Bovine Serum (FBS, 16140071, Invitrogen, ThermoFisher, USA), LS174T cells were culture in MEM + 10% FBS + 1% P/S (SU 17920260, HaoranBio, China), while HT29 and HCT116 cells were cultured in McCoy’s 5A (12330031, Gibco, ThermoFisher, USA) containing 10% FBS and 1% P/S, and HTC8 and HCT-15 cells were cultured in RPMI-1640 (61870044, Gibco, ThermFisher, USA) containing 10% FBS + 1% P/S. Moreover, SW-620 cells were cultured in DMEM high-glucose (10569010, Gibco, ThermoFisher, USA) containing 10% FBS, 1% P/S, and LOVO cells were cultured in 95% MEM with 5% DMSO (D12345, Gibco, ThermoFisher, USA).
Immunohistochemistry
FFPE samples (CRC and adjacent tissues) were sectioned into 4µm thick, and prepared for immunohistochemistry staining. The sections were incubated with rabbit polyclonal anti-FAT4 antibody (ab243591, 1:200, Abcam, UK) at 4 °C overnight. After washing the sections by PBS, the sections were then incubated with goat anti-rabbit secondary antibody (ab205718, 1:2,000, Abcam, UK) at room temperature for 30 min. The peroxidase reactivity was measured by 3,3’-diaminobenzidine (DAB) (7411-49-6, Suzhou Yacoo Chemical Reagent Co., Ltd, China) and counterstained by haematoxylin. The IHC evaluation was conducted according to the description of Kusinska (16). The score grade of IHC intensity was classified as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). Based on the scores, positive cells were accordingly assigned into four groups, namely, 0 (negative), 1(less than 10%), 2 (from 10% to 50%), and 3 (more than 50%) groups. The score of IHC intensity multiplied by the percentage of positive cells was expressed as the final score. If the final score was lower than or equal to 1, it was considered to be negative, while the score ranging from 2 to 9 was positive.
Transfections
SiFAT4 (SR312478) and SR312478-FAT4 overexpression plasmid were obtained from Origene, siRNA (A01001) was purchased from Gene pharma. LS174T and SW-620 cell lines were cultured in 6-well plates (5×105 cells per well) to reach confluence of 60% to 80%. Lipofectamine TM 2000 transfection reagent (#11668019, ThermoFisher, USA) was used according to its instruction. A total volume of 75 µg siFAT4 or siRNA was prepared and added to LS174T cells, while FAT4 and an empty plasmid was respectively added to SW-620 cells, and the cell lines were all cultured in DMEM medium. After culturing the cells in CO2 for 6 h at 37 °C, DMEM medium was replaced by 10% FBS/DMEM medium to continue the culture for another 24 h.
Quantitative real time PCR
The expression levels of FAT4 and EMT-related genes were measured by qRT-PCR. Total RNAs were extracted from FFPE tissues or cell samples by PureLink™ RNA Mini Kit (12183018A, ThermoFisher, USA). RNA concentration was determined by NanoDrop 2000c (# ND2000CLAPTOP, Thermo Fisher, USA). Reverse transcription from RNAs to cDNAs was carried out using miScript Reverse Transcription Kit (218160, Qiagen GmbH, German) following the instructions. SYBR-Green PCR kit and the Fast Real-Time PCR system (ABI 7500, Applied Biosystems, Carlsbad, CA) were used for qRT-PCR.
The thermocycling conditions were set as follows: at 95 °C for 20 min, and then 40 cycles at 94 °C for 15 s, at 55 °C for 30 s and at 72 °C for 30 s. 2-ΔΔCT was used to measure the expression levels of FAT4 and EMT-related biomarkers (17). All primer sequences were listed in Table 1.
Table 1
| Gene | Primer | Sequence |
|---|---|---|
| FAT4 | Forward | 5' TTGAAGGAAGGAGAACCCAT3' |
| Reverse | 5' TCGTCCAATAGTAAAGAGGC3' | |
| VEGF | Forward | 5' GAGGAGCAGTTACGGTCTGTG3' |
| Reverse | 5' TCCTTTCCTTAGCTGACACTTGT3' | |
| E-Cadherin | Forward | 5' CGAGAGCTACACGTTCACGG3' |
| Reverse | 5' GGGTGTCGAGGGAAAAATAGG3' | |
| N-Cadherin | Forward | 5' TTTGATGGAGGTCTCCTAACACC3' |
| Reverse | 5' ACGTTTAACACGTTGGAAATGTG3' | |
| Vimentin | Forward | 5' GCCCTAGACGAACTGGGTC 3' |
| Reverse | 5' GGCTGCAACTGCCTAATGAG 3' | |
| GADPH | Forward | 5' CGGAGTCAACGGATTTGGTCGTAT3' |
| Reverse | 5' AGCCTTCTCCATGGTGGTGAAGAC3' |
Group separation
FAT4 expressions in cell lines treated by 5-FU (T0984, Top Science, USA) at different concentrations (0, 1, 5, 10, 30, 60, 120 µg/mL) at 37 °C for 24 and 48 h were determined, so as to identify the cell lines with the highest FAT4 expression (LS174T) and the lowest (SW-620). The optimal concentration of 5-FU was defined as 30 µg/mL. The cells were divided into four groups, namely, LS174T: siNC, siFAT4, siNC+5-FU (30 µg/mL), siFAT4+5-FU. SW-620: mock, FAT4, mock + 5-FU, FAT4 + 5-FU.
Western blot
The levels of FAT4-related proteins and EMT-related proteins were measured by Western blot. RIPA lysis buffer solution was prepared for the lysates of cell lines. After 20-min reaction on ice, the solutions were centrifuged to collect supernatants.
BCA protein assay (Bio-Rad, Hercules, CA, USA) was used to measure the protein concentration. Proteins (30 µL) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). Skimmed milk (5%) was used to seal the membranes for 2 h at room temperature, then the membranes were incubated with specific antibody for FAT4 (anti-rabbit, Abcam, ab130076, 1:500, UK), VEGF (anti-mouse, Abcam, ab1316, 1:100, UK), E-Cadherin (anti-rabbit, Abcam, ab40772, 1:10,000, UK), N-Cadherin (anti-rabbit, Abcam, ab18203, 1:1,000, UK), Vimentin (anti-rabbit, Abcam, ab92547, 1:1,000, UK) at 4 °C overnight. TBST was used to wash the membranes for 3 times, and then the membranes were incubated with the respective secondary antibody (goat anti-rabbit, Abcam, ab150077, 1:1,000, UK, goat anti-mouse, Abcam, ab150113, 1:200, UK) at room temperature for 2 h, and washed by TBST for 3 times and developed by ECL prime Western blot (BRPN2232, Sinopharm Chemical Reagent). The bands were conventionally pressed and photographed. The optical density was analyzed by Image J (Image J 1.8.0, National Institute of Health). GAPDH (anti-mouse, Abcam, ab8245, 1:500, UK) served as the internal references.
Cell counting kit-8 assay (CCK-8)
The cell viabilities of LS174T, SW-620 cell lines and siNC, siFAT4, siNC + 5-FU, siFAT4 + 5-FU, mock, FAT4, mock + 5-FU, FAT4 + 5-FU were detected by CCK-8 assay. The cells were seeded in 96-well plates, and stained using CCK-8 solution (HY-K0301, MedChemExpress, USA) at 37 °C for 2 h. Optical density (OD) value was measured at the wavelength of 450 nm.
Flow cytometry assay
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection Kit (70-AP101-100-AVF, MultiSciences, China) was applied to detect cell apoptosis. The cells were separated by 0.25% trypsin (25300054, Gibco, Thermofisher, USA), and cell solution was prepared at the density of 106/mL. After centrifugation, the sediments were collected. A dose of 2 mL PBS was added into 1mL cells and centrifuged again to collect the sediments. The sediments were then fixed by 70% ethanol at 4 °C for 2 h, then rinsed in PBS twice. After centrifugation, the sediments were resuspended in binding solution. The reaction was conducted using 10 µL Annexin V-FITC and 5 µL PI for 15 min, then in another 300 µL binding buffer in the dark. Then cell apoptosis was measured by flow cytometry (NJ 08855-1327, BD FACSCanto II, BD Biosciences, Franklin Lakes, USA).
Colony formation assay
After cell digestion, the solution was centrifuged, the sediments were collected and cultured in a 6-well plate at the density of 10×103 per well for 10 days. Then cells were rinsed in PBS buffer, fixed by 1 mL methanol for 15 min at room temperature and stained by crystal violet solution (0.1%) for 20 min. Finally, the cells were finally photographed, counted and analyzed.
Migration and invasion assays
Cell migration was measured by wound-healing assays. 1.5×105 LS174T or SW-620 cells (1.5×105 cells/well) were respectively seeded into a 12-well culture plate. After transfection of each cell lines for 24 h, the cells were scratched using a sterilized micropipette. The medium was replaced by serum-free and the wound was photographed at 0 and 24 h using Image J software (NIH Image, Bethesda, MD).
The invasion abilities of LS174T and SW-620 cell lines were detected by transwell assays. A 12-well Transwell cell culture chamber (8 µm, 430172, corning Costar Crop., USA) was used, and all the procedures were conducted following the manufacturer’s instructions. After the transfection for 24 h, the cells were suspended in the upper chambers at a density of 4×104 cells/mL on 50-µL Matrigel (2.0 mg/mL, 354234, Corning, USA) layer. A total of 500 µL DMEM medium containing 10% FBS was seen as the lower chemoattractant. The cells were incubated with 5% CO2 at 37 °C for 24 h. The cells in the upper chambers were stained by crystal violet and cell invasion capacity was measured under a microscope.
Tube formation assay
Ibidi angiogenesis slide (Ibidi, Martinsried, Germany) was used for tube formation measurement. Matrigel was stored at 4 °C overnight in advance, and 10 µL Matrigel was then added into each well until coagulated. A total of 50 µL cell suspension (2×105 cells/mL) was then respectively added into each well, while cell-free culture medium was added into upper well. The images were regularly observed, and coverage area and cell formation number were collected for statistical analysis.
Statistical analysis
The data were analyzed by SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) and shown as mean ± standard deviation. Student’s t-test (two-tailed) and one-way ANOVA followed by Dunnett’s post hoc test were carried out for statistical analyses. All the experiment was conducted in triplicate. P<0.05 was considered to be a statistically significant difference
Results
FAT4 expression in CRC tissues and cells
Immunohistochemistry and qRT-PCR were conducted to detect FAT4 expressions in CRC and adjacent tissues. Immunohistochemistry results showed a notable FAT4 staining in adjacent tissues in comparison with a significantly less staining strength in CRC tissues (Figure 1A). The result from qRT-PCR detection showed that the FAT4 expression was greatly higher in adjacent tissues than in CRC cancer tissues (Figure 1B).
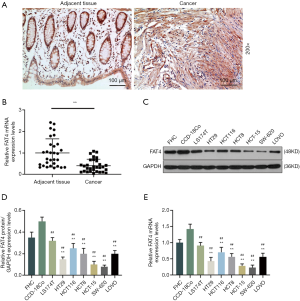
QRT-PCR and Western blot were performed to determine gene and protein expressions of FAT4 in normal colon cell line, normal colorectal mucosa cell line and CRC cell lines. Compared to FHC cell line, HT29, HCT116, HCT8, HCT-15, SW-620 and LOVO cell lines had a significantly lower gene and relative protein levels of FAT4 (P<0.001, Figure 1C,D,E), while CCD-18Co cell line had the highest FAT4 and relative protein expressions in all the cell lines, showing a significant difference from CCD-18Co and LS174T, HT29, HCT116, HCT8, HCT-15, SW-620 and LOVO cell lines.
The effect of different 5-FU doses on cell viability and FAT4 expression of LS174T and SW-620 cells
According to CCK-8 results, the OD value of LS174T cells was reduced as the concentrations of 5-FU increased, and a significant difference appeared when the 5-FU concentration reached 5 µg/mL and higher (Figure 2A). The OD value of LS174T cells was higher at 48 h than at 24 h, when the 5-FU concentration was 60 µg/mL and lower. In contrast, the OD value on 48 h was lower than at 24 h when 5-FU concentration was used at 120 µg/mL. The similar result was observed on SW-620 cells (Figure 2B), the increases in 5-FU concentrations significantly reduced the OD value of SW-620 cells at 24 and 48 h, moreover, the OD values of SW-620 at 24 h was higher than that at 48 h with any 5-FU concentration.
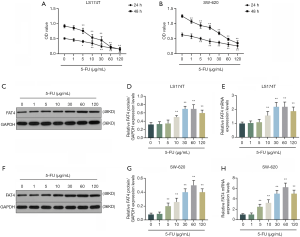
QRT-PCR and Western blot results showed that 5-FU at 10 µg/mL or higher could significantly up-regulate the FAT4 expressions in LS174T and SW-620 cells (Figure 2C,D,E,F,G,H). The FAT4 expressions kept increasing and reached the highest when the 5-FU concentration was 60 µg/mL, and then the expression decreased at 120 µg/mL 5-FU.
FAT4 transfection efficiency in LS174T and SW-620 cells
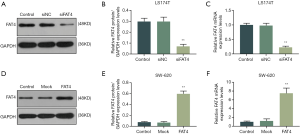
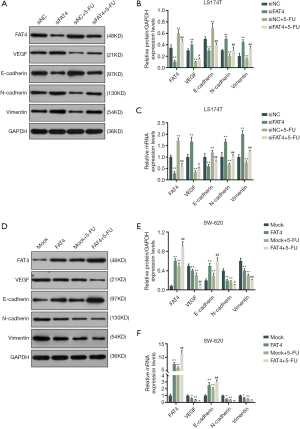
The transfection efficiency of FAT4 in LS174T and SW-620 cells were detected by qRT-PCR and Western blot (Figure 3). In LS174T cells, the expression of FAT4 was silenced, and results showed that both relative mRNA and protein expressions were significantly reduced (P<0.001). In comparison of SW-620 cells transfected with overexpress FAT4 plasmid, the expression levels of the mRNA and protein were increased (P<0.001).
Effects of FAT4 on cell sensitivity to 5-FU
The cell viabilities of LS174T and S-620 cell lines were detected by CCK-8 assay (Figure 4A,B). In LS17T cell line, silencing FAT4 expression could significantly increase the cell OD values at 24 h and 48 h, whereas in SW-620 cells, overexpression of FAT4 had a lower OD value with or without 5-FU addition. Moreover, the OD value of both LS174T and SW-620 cell lines decreased with the addition of 5-FU.
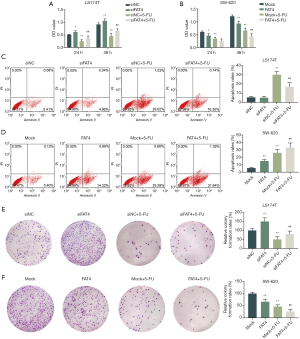
Apoptosis of LS174T and SW-620 was determined (Figure 4C,D), and the results showed that the silence of FAT4 in LS174T had no significant effect on cell apoptosis rate, however, when FAT4 was silenced in LS174T pretreated by 5-FU, the apoptosis was significantly reduced compared to the cells only treated by 5-FU. Furthermore, the result of SW-620 showed that FAT4 overexpression significantly up-regulated cell apoptosis rate. Moreover, the addition of 5-FU in the two cell lines could significantly increase the cell apoptosis.
The colony formation results showed that silencing FAT4 significantly up-regulated the cell colony formation rate, which could be greatly reduced by overexpression of FAT4 and the addition of 5-FU (Figure 4E,F).
The cell migration rate was determined, and results showed that LS174T cell migration rete was significantly increased with the silence of FAT4, however, the overexpression of FAT4 greatly reduced the cell migration rate of SW-620. Additionally, the 5-FU treatment noticeably lowered the migration rates of the two cell lines (Figure 5A,B).
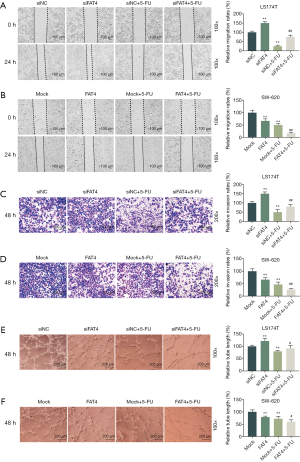
Similarly, the cell invasion rates were increased significantly by silencing of FAT4, but were decreased significantly when FAT4 was overexpressed. The pretreatment with 5-FU could significantly reduce the cell invasion rates (Figure 5C,D).
The tube formation was measured, and results showed that the silence of FAT4 could significantly increase the tube length, while the FAT4 overexpression resulted in a significant shorter tube length (Figure 5E,F).
The effect of moderating FAT4 expression on EMT process in LS174T and SW-620 cells
The protein and mRNA expressions of FAT4 and EMT were measured by qRT-PCR and Western blot (Figure 6). In LS174T cells, the silencing of FAT4 decreased the relative protein and mRNA expressions of FAT4 and E-Cadherin, and increased the expressions of VEGF, N-Cadherin, Vimentin. However, increasing FAT4 expression up-regulated the expressions of FAT4 and E-cadherin and down-regulated the relative protein and mRNA expressions of VEGH, N-cadherin and Vimentin. Moreover, the 5-FU treatment could increase the FAT4 and E-cadherin expressions, and reduce the expressions of VEGF, N-cadherin and Vimentin.
Discussion
FAT gene family is a newly discovered gene member, and has been widely reported to have biological functions (18-20). For the functions of FAT4 in tumors, Sun investigated the relation between FAT4 and SOX11 in gastric cancer, and found that abnormal methylation of both can largely increase the risk developing suffering gastric cancer (21). Moreover, 30% invasive lobular carcinoma showed FAT4 mutation (22). In the present study, FAT4 showed a significant lower expression in CRC cancer tissues than adjacent tissues (P<0.001), indicating a potential role of FAT4 in CRC. Moreover, the prognosis of CRC is likely to be related to FAT expression. Previous study investigated the relation between prognosis of gastric cancer and FAT4 expressions, and found that high FAT4 promoter methylation was related to a knockdown of FAT4 expression, thus resulted in a poor prognosis of gastric cancer (23). The survival rate of breast cancer was also reported to be associated with FAT4 hippo signaling pathway, as higher FAT4 expression greatly increases the potential of a patient having better prognosis (24). In our research, LS174T had the highest FAT4 expression, while SW-620 had the lowest FAT4 expression in all the cell lines.
The effects of 5-FU on cell viability were investigated in the present study, and the results showed that the cell viability of SW-620 was reduced more significantly at lower 5-FU dose, indicating that SW-620 cells were more sensitive to 5-FU than LS174T. On the other hand, once the concentration of 5-FU reached higher than 5µg/mL, the cell viability could be significantly affected, moreover, 5-FU showed great effects on FAT4 expressions in LS174T and SW-620, as the expression of FAT4 was greatly increased in the two cells by the pretreatment of 5-FU. However, the FAT4 expression was significantly inhibited when the concentration of 5-FU reached 120 µg/mL (P<0.001), indicating the potential negative effect resulted from high 5-FU dose. Kobayashi et al. has also found similar relation between high 5-FU concentration and prognosis of pancreatic cancer patients, as they observed that a high 5-FU concentration caused poor prognosis in cancer pancreatic patients (25).
The expression of FAT4 in LS174T and SW-620 cells was determined when FAT4 was silenced or overexpressed. In the cells with high FAT4 expression, silencing of FAT4 expression could significantly lower FAT4 expression (P<0.001). However, SW-620 first showed a rather low FAT4 expression, but after transfection with FAT4 overexpression plasmid, the up-regulated FAT4 was observed in SW-620 cells (P<0.001). FAT has been reported to control organ size by regulating cell cycles (26,27). Research showed that the block of FAT4 expression could increase tumor size, and results in more aggressive invasion and migration of tumor cells (28). Moreover, silencing FAT4 expression significantly activated cell viability, reduced cell apoptosis rate and increased colony formation, cell migration and invasion rates (P<0.001), suggesting that FAT4 could strongly inhibit CRC cell activity. Moreover, tube formation plays a critical role in the process of tumor development and metastasis, in the present study, tube formation was longer after the knockdown of FAT4 expression, indicating that FAT4 inhibition was possibly related to tumor metastasis. However, the FAT4 overexpression results indicated that the presence of FAT4 could suppress tumor metastasis. The addition of 5-FU also showed a similar results. Thus, the data suggested that FAT4 could promote the effects of 5-FU on suppressing cancer cell activity on cell proliferation, migration, and promoting CRC cell apoptosis. It is reported that down-regulation of Hippo pathway was able to attenuate the resistance of CRC cells to 5-FU (29). As explained above, FAT4 acts as a mediator in Salvador-Warts-Hippo signaling pathway, thus, FAT4 might play a role in the correspondence between 5-FU and FAT4.
E-Cadherin and N-Cadherin are EMT markers, apart from the two, VEGF and Vimentin are also commonly considered to be EMT markers (30,31). Gravdal reported that high level of N-Cadherin and low level of E-Cadherin was strongly associated with death caused by prostate cancer (32). Cai et al. reported that FAT4 played a tumor suppressor role in gastric cancer by inhibiting EMT (33). In the present research, the expression levels of VEGF, E-Cadherin, N-Cadherin and Vimentin and the FAT4 expression were determined, the results showed that E-cadherin was low-expressed, but VEGF, N-Cadherin, and Vimentin were high-expressed in case that FAT4 was silenced. However, the high E-Cadherin and low VEGF, N-Cadherin, Vimentin expression levels were observed after overexpressing FAT4. Such results suggested a possible connection between FAT4 and EMT process, specifically, FAT4 was likely to increase the expression levels of E-Cadherin but suppress the expressions of VEGF, N-Cadherin, Vimentin, thus inhibiting the EMT process.
Conclusions
In conclusion, the present study finds the relationship between FAT4 and CRC, specifically, FAT4 functions as a tumor suppressor and has the ability to enhance the 5-FU effect on suppressing the growth, migration, invasion, and tube formation of CRC cells, in other words, CRC cells are more sensitive to 5-FU with overexpressed FAT4. Furthermore, FAT4 is likely to inhibit EMT of CRC cells by up-regulating the expression of E-Cadherin and down-regulating the expressions of VEGF, N-Cadherin and Vimentin.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.74). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All patients and their relatives were informed and signed the consent form for allowing their tissues to be used in the research. The study was approved by the First Affiliated Hospital of Zhejiang Chinese Medical University Ethics Committee (2019).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fedewa SA. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Ndreshkjana B, Capci A, Klein V. Combination of 5-fluorouracil and thymoquinone targets stem cell gene signature in colorectal cancer cells. Cell Death Dis 2019;10:379. [Crossref] [PubMed]
- McQuade RM, Stojanovska V, Bornstein JC. Colorectal Cancer Chemotherapy: The Evolution of Treatment and New Approaches. Curr Med Chem 2017;24:1537-57. [Crossref] [PubMed]
- Tang JC, Feng YL, Liang X. Autophagy in 5-Fluorouracil Therapy in Gastrointestinal Cancer: Trends and Challenges. Chin Med J (Engl) 2016;129:456-63. [Crossref] [PubMed]
- Zhao P, Ma YG, Zhao Y. MicroRNA-552 deficiency mediates 5-fluorouracil resistance by targeting SMAD2 signaling in DNA-mismatch-repair-deficient colorectal cancer. Cancer Chemother Pharmacol 2019;84:427-39. [Crossref] [PubMed]
- Zhang X, Pang W, Liu H. Lidocine potentiates the cytotoxicity of 5-fluorouracil to choriocarcinoma cells by downregulating ABC transport proteins expression. J Cell Biochem 2019;120:16533-42. [PubMed]
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol 2009;41:349-69. [Crossref] [PubMed]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 2006;16:2101-10. [Crossref] [PubMed]
- Dent LG, Poon CL, Zhang X. The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway. Curr Biol 2015;25:124-30. [Crossref] [PubMed]
- Qi C, Zhu YT, Hu L. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer 2009;124:793-8. [Crossref] [PubMed]
- Eldeeb MA, Fahlman RP, Esmaili M, Ragheb MA. Regulating Apoptosis by Degradation: The N-End Rule-Mediated Regulation of Apoptotic Proteolytic Fragments in Mammalian Cells. Int J Mol Sci 2018; [Crossref] [PubMed]
- Han Y, Jin X, Li H. Microarray analysis of copy-number variations and gene expression profiles in prostate cancer. Medicine 2017;96:e7264. [Crossref] [PubMed]
- Zhou D, Kannappan V, Chen X. RBP2 induces stem-like cancer cells by promoting EMT and is a prognostic marker for renal cell carcinoma. Exp Mol Med 2016;48:e238. [Crossref] [PubMed]
- Li Y, Su X, Pan H. Inhibition of lncRNA PANDAR reduces cell proliferation, cell invasion and suppresses EMT pathway in breast cancer. Cancer Biomark 2019;25:185-92. [Crossref] [PubMed]
- Matsubara T, Tagawa T, Takada K. Clinical and Prognostic Significance of the Epithelial-Mesenchymal Transition in Stage IA Lung Adenocarcinoma: A Propensity Score-Matched Analysis. Clin Lung Cancer 2019;20:e504-13. [Crossref] [PubMed]
- Kusinska RU, Kordek R, Pluciennik E. Does vimentin help to delineate the so-called 'basal type breast cancer'? J Exp Clin Cancer Res 2009;28:118. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Zhang X, Liu J, Liang X, Chen J. History and progression of Fat cadherins in health and disease. Onco Targets Ther 2016;9:7337-43. [Crossref] [PubMed]
- Dogan S, Xu B, Middha S. Identification of Prognostic Molecular Biomarkers in 157 HPV-positive and HPV-negative Squamous Cell Carcinomas of the Oropharynx. Int J Cancer 2019;145:3152-62. [Crossref] [PubMed]
- Cheng H, Burroughs-Garcia J, Birkness JE. Disparate Regulatory Mechanisms Control Fat3 and P75NTR Protein Transport through a Conserved Kif5-Interaction Domain. PloS One 2016;11:e0165519. [Crossref] [PubMed]
- Badouel C, Zander MA, Liscio N, et al. Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development 2015;142:2781-91. [Crossref] [PubMed]
- Ping Z, Siegal GP, Harada S. ERBB2 mutation is associated with a worse prognosis in patients with CDH1 altered invasive lobular cancer of the breast. Oncotarget 2016;7:80655-63. [Crossref] [PubMed]
- Jiang X, Liu Z, Xia Y. Low FAT4 expression is associated with a poor prognosis in gastric cancer patients. Oncotarget 2017;9:5137-54. [PubMed]
- Bliss SA, Paul S, Pobiarzyn PW. Evaluation of a developmental hierarchy for breast cancer cells to assess risk-based patient selection for targeted treatment. Sci Rep 2018;8:367. [Crossref] [PubMed]
- Kobayashi S, Ueno M, Omae K. Influence of initial dose intensity on efficacy of FOLFIRINOX in patients with advanced pancreatic cancer. Oncotarget 2019;10:1775-84. [Crossref] [PubMed]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer 2007;7:182-91. [Crossref] [PubMed]
- Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol 2009;1:a003129. [Crossref] [PubMed]
- Mao Y, Kuta A, Crespo-Enriquez I. Dchs1-Fat4 regulation of polarized cell behaviours during skeletal morphogenesis. Nat Commun 2016;7:11469. [Crossref] [PubMed]
- Sebio A, Matsusaka S, Zhang W. Germline polymorphisms in genes involved in the Hippo pathway as recurrence biomarkers in stages II/III colon cancer. Pharmacogenomics J 2016;16:312-9. [Crossref] [PubMed]
- Vuoriluoto K, Haugen H, Kiviluoto S. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 2011;30:1436-48. [Crossref] [PubMed]
- Yang HL, Thiyagarajan V, Shen PC. Anti-EMT properties of CoQ0 attributed to PI3K/AKT/NFKB/MMP-9 signaling pathway through ROS-mediated apoptosis. J Exp Clin Cancer Res 2019;38:186. [Crossref] [PubMed]
- Gravdal K, Halvorsen OJ, Haukaas SA. A switch from E-cadherin to N-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin Cancer Res 2007;13:7003-11. [Crossref] [PubMed]
- Cai J, Feng D, Hu L. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/beta-catenin signalling. Br J Cancer 2015;113:1720-9. [Crossref] [PubMed]


