
Pediatric cerebellar tumors: transcriptionally distinct but developmentally heterogeneous
The cerebellum is composed of a wide variety of cell types. Many of these cell types undergo temporally regulated differentiation through different developmental stages (1). The cerebellar neurons that use γ-aminobutyric acid (GABA) or glutamate as neurotransmitters, differentially arise from two spatially distinct germinal zones of the cerebellar anlage: the cerebellar ventricular zone (CVZ), characterized by progenitor cells expressing Ptf1a; and rhombic lip (RL), characterized by progenitor cells expressing Atoh1 (Figure 1A) (2). Cerebellar neurons along with cerebellar glial cells, migrate into precise, spatially organized regions in the cerebellum during development of cerebellum. Ultimately, adult cerebellar neurons develop into either inhibitory gamma butyric acid (GABAergic) or excitatory glutamatergic types (2,3).
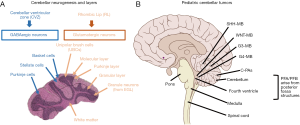
Pediatric brain tumors most frequently arise from the posterior fossa of the cerebellum. The most prevalent pediatric cerebellar tumors are medulloblastoma, ependymoma, and cerebellar pilocytic astrocytoma (C-PA) (Figure 1B). Medulloblastoma tumors show well-characterized intertumoral heterogeneity, as well as spatial (in metastases) and temporal heterogeneity at recurrence (4-7). Medulloblastoma is conventionally classified into four molecular subgroups: WNT, SHH, Group 3, and Group 4. Ependymomas exhibit marked intertumoral heterogeneity (4,8-10) and can arise throughout the central nervous system (8,11). Transcriptional profiling has demonstrated the existence of two demographically, transcriptionally, genetically, and clinically distinct subtypes: Group A posterior fossa ependymoma (PFA), patients are younger, possess laterally located tumors, and more likely to exhibit recurrence with metastasis; PFA Group B patients (PFB) (9,10). Genome-wide methylation profiling also sub-classified PFA into Group A, increased DNA methylation in CpG islands, and Group B, less genome wide methylation (12). C-PAs are not well characterized with regards to intertumoral heterogeneity (13). Currently pediatric cerebellar tumors are treated with non-specific therapies, which contributes to poor patient response and neurotoxicity. To better understand the biology of cerebellar tumors it is important to identify and study their cell of origin and development.
Recently, there has been significant progress in understanding the developmental origins of the cerebellum, as a result of molecular profiling using single cell RNA sequencing or scRNAseq. Recent studies have shown that clustering of the cells of the presumptive GABAergic lineage delineate from nine cell groups (14). The study of the origin and development of human cerebellar tumors has been more challenging, but progress has been made on this front as well. Vladoiu et al. (Nature, 2019) utilized scRNAseq to define the cell of origin of three of the four medulloblastoma molecular subgroups (SHH, Group 3, Group 4) (6,7); ependymomas PFA/PFB; and C-PA (15). The authors analyzed cells from various stages of the developing normal mouse cerebellum and compared these to human pediatric cerebellar tumors. The authors report that these different childhood tumors mirror the transcription of cells from different temporally defined cerebella cell lineages. Specifically, SHH-MB is best matched to the granule cell progenitor (GCP) lineage in the early postnatal period, while Group 4-MB, PFA and PFB, and C-PA matches best to the cell clusters observed during fetal development. However, a striking finding is that the different medulloblastoma subgroups along with PFA and C-PAs, possess extensive single cell heterogeneity with evidence cells from different lineages of differentiation (15).
Although PFA/PFAB ependymomas and cerebellar C-PAs are both pediatric cerebellar tumors, transcriptome analysis mirrors this in terms of gliogenesis. Interestingly, this study demonstrated that expression of nestin is seen primarily in early development, whereas glutamatergic and GABAergic populations of neurons appeared in the mid-cerebellar developmental stage, and glial cells develop at a later stage.
According to the authors, non-transformed cell populations that transcriptomically match closest to the human cerebellar tumors are only found in utero or immediate postnatal stages. The results of this study indicate that these cerebellar tumors develop from a disorganization of early brain development. This explains the enigma that these cell populations are not present in the brain of children with cerebellar tumors at diagnosis/presentation.
Single cell RNAseq (scRNAseq) data with unsupervised clustering of cell transcriptomes generated more than 30 individual clusters in this study. Majority of these clusters are heavily populated with cells from specific developmental time points. Cerebellar samples of five embryonic time points and four early postnatal time points from wild-type mice were identified. On the basis of established single-cell-type transcriptional profiles of mouse, the authors tried to match the transcriptomes of bulk human ependymomas (PFA and PFB) and cerebellar C-PAs to distinct cell clusters of specific cerebellar developmental stages.
Medulloblastomas (SHH, Group 3, Group 4)
The authors were able to pinpoint the cellular origins of medulloblastomas. SHH-MB cells appear to overall mirror the granule cell hierarchy. By comparing the bulk human SHH-MB transcriptome against expression signatures of granule progenitors the authors identified seven granule cell sub-clusters. The prognostic importance of this finding is that they found SHH-MB that resemble later time points in cerebellar development and characterized as the SHH1, have a worse prognosis than the SHH2 group which resemble early arising granular cell precursors. Recent work has identified four clinically and cytogenetically distinct groups of SHH-MB: SHHα, SHHβ, SHHγ, and SHHδ (5). Transcriptome of SHHβ medulloblastomas which represents the SHH-MB observed in infants matches best with that of the SHH-2 subset, demonstrating that this SHH2 subtype is developmentally connected to differentiation states in early GCP development.
The authors could not categorize the exact developmental origin of Group 3-MBs using bulk transcriptomics, since their study revealed that Group 3-MB often transcriptionally matches to more than one cluster in the developing cerebellum. Analysis of Group 3-MB cell clusters showed that it’s transcriptome shares similarity with Nestin + cerebellar early stem cells. However, the authors found a subset of bulk Group 3-MB transcriptome that matches developmental cell clusters in GCP and UBC lineages and other subsets showing similarity to GABAergic interneurons. The authors intriguingly found transcriptome analysis of single cells, revealed clusters of progenitors belonging to glutamatergic (Atoh1), and GABAergic (Ptf1a) neurons. Stem cell-like clusters expressing Nestin was observed in early time points of neural cell development, whereas glutamatergic and GABAergic neuronal populations appeared in mid-development, and the glial cells developed at a later stage of development. In contrast, the non-CNS cells were found across all time points. By transcriptional mapping of individual tumor cell clusters from Group 3-MB the authors demonstrated the existence of highly divergent lines of differentiation, in tumor clusters similar to a number of normal developmental clusters in the GCP, UBC, Purkinje cell and GABAergic interneuron lineages. This pattern of similarity in transcriptional lineage opened up new insights that Group 3-MB arises from an early, uncommitted cerebellar stem cell, followed by partial differentiation of transformed cells. The source of origin in Group 3-MB is not definitively known and suggested to be Nestin+ cerebellar stem cell. A rare population of Nestin expressing precursors (NEPs) has been already identified in mouse cerebellum (16). Unlike conventional granule neuron progenitors (GNPs), which are found in the outer external granular layer (EGL) and proliferate substantially, NEPs accumulate in the deeper part of the EGL and remain quiescent (16). The study by Vladoiu et al. detected that a subset of bulk Group 3-MB transcriptomes resemble developmental cell clusters in the GCP and UBC lineages. The UBC and GCP progenitors are similar to cerebellar GABAergic interneurons and this multi-lineage differentiation corroborates a model in which Group 3-MBs originates from early neural stem-like cells which are Nestin positive and give rise to a variety of differentiated progeny.
scRNAseq of human Group 4-MB mirror the transcriptome of both differentiated and progenitor unipolar brush cells (UBCs). However, some Group 4-MB tumor tissues revealed a transcriptome similar to GCP lineage. So far, the cell of origin of Group 4-MB was not definitely known and mouse model of Group 4-MB is unavailable. One of the most significant finding is that these cells of UBC lineage are glutamatergic interneurons develop from the upper RL of the cerebellum. Thus, this cluster represents a well-defined source of Group 4-MB and is first observed in mouse cerebellum E14 peaks and highly prevalent in embryonic day 18, disappearing by P0. Their data confirmed for the first time that the cell of origin of human Group 4-MB is a unique group of UBC and GCP progenitor cluster that simultaneously express GCP and UBC marker genes. Their data identifies that human Group 4-MB arise from bi-potential progenitor cell populations which possess the ability to give rise to cells in both GCP and UBC lineages. Finally, their work revealed a cell in the UBC lineage from upper RL of cerebellum is the source of origin for the highly aggressive Group 4-MB.
PFA (PFA and PFB) and C-PAs
Comparison of the transcriptomic analysis of bulk ependymomas with C-PAs showed that C-PAs are not only clinically and histologically different, but have different cells of origin. Specifically, C-PAs arise from distinct cerebellar gliogenic progenitor cells which are different from gliogenic progenitor cells from early ventricular radial glia and roof-plate stem cells (source of origin for PFA and PFB). The data presented in this article show that a substantial proportion of differentiated cells within PFA or differentiated glioma in MBs do not match a majority of differentiated neuronal cells within cerebellar C-PAs. Interestingly, the normal non-transformed cell populations that match transcriptionally with cerebellar tumors found only in embryonic or early post-natal stage were not found in children at disease presentation. The authors pointed that when bulk PFA and PFB transcriptomes were compared to mouse single cells in the gliogenic progenitor cell lineage from developmental stages through E10 to P0, a specific match was observed with the E16 gliogenic progenitors. This study revealed an absence of differentiated cell types only in PFA, but not in other cerebellar tumor types, indicating the prevalence of a differentiation block in PFA.
Conclusions
The authors acknowledge that it would have been of greater value to compare single-cell transcriptome of human cerebellar tumors with normal human cerebellar cells from different cerebellar developmental time points, since the embryonic and postnatal time periods are different in humans and mice. Still, this study provides a detailed insight of the biology and transcriptomes of the specific cerebellar hierarchies and how that relates to the development of different types of human cerebellar tumors. This work should accelerate development of new mouse models of pediatric cerebellar brain tumors and potentially hasten development of new therapies.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xian-Xin Qiu (Shanghai Proton and Heavy Ion Center (SPHIC), a.k.a. the Proton and Heavy Ion Center of Fudan University Shanghai Cancer Center (FUSCC), Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.31). DPK declares being on a patent application (19109 PCT) filed from Emory University which is currently in pending status. SS reports that she has provided consulting support to Novocure, as an advisory committee member once and has received an income from Novocure for consultation provided. In addition to this Dr. Sengupta has patent 19109 PCT filed from Emory University in pending status. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci 1998;21:375-82. [Crossref] [PubMed]
- Carter RA, Bihannic L, Rosencrance C, et al. A Single-Cell Transcriptional Atlas of the Developing Murine Cerebellum. Curr Biol 2018;28:2910-20 e2.
- Carletti B, Rossi F. Neurogenesis in the cerebellum. Neuroscientist 2008;14:91-100. [Crossref] [PubMed]
- Morrissy AS, Cavalli FMG, Remke M, et al. Spatial heterogeneity in medulloblastoma. Nat Genet 2017;49:780-8. [Crossref] [PubMed]
- Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017;31:737-54.e6. [Crossref] [PubMed]
- Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311-7. [Crossref] [PubMed]
- Northcott PA, Shih DJ, Peacock J, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012;488:49-56. [Crossref] [PubMed]
- Pajtler KW, Witt H, Sill M, et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015;27:728-43. [Crossref] [PubMed]
- Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 2011;20:143-57. [Crossref] [PubMed]
- Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 2005;8:323-35. [Crossref] [PubMed]
- Pajtler KW, Mack SC, Ramaswamy V, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol 2017;133:5-12. [Crossref] [PubMed]
- Fukuoka K, Kanemura Y, Shofuda T, et al. Significance of molecular classification of ependymomas: C11orf95-RELA fusion-negative supratentorial ependymomas are a heterogeneous group of tumors. Acta Neuropathol Commun 2018;6:134. [Crossref] [PubMed]
- Jones DT, Hutter B, Jager N, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 2013;45:927-32. [Crossref] [PubMed]
- Wizeman JW, Guo Q, Wilion EM, et al. Specification of diverse cell types during early neurogenesis of the mouse cerebellum. Elife 2019; [Crossref] [PubMed]
- Vladoiu MC, El-Hamamy I, Donovan LK, et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 2019;572:67-73. [Crossref] [PubMed]
- Li P, Du F, Yuelling LW, et al. A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat Neurosci 2013;16:1737-44. [Crossref] [PubMed]


