
Comprehensive analysis reveals a four-gene signature in colorectal cancer
Introduction
Colorectal cancer (CRC) is known as one of the most common non-skin cancers diagnosed both in men and women in the world. CRC often begins as a polyp inside the colon or rectum. Adenomas, a form of polyps, are innocent tumors within the tissue of the colon or rectum (1). Though most polyps will stay benign, some of them have the probabilities of converting into cancer as time goes on. There were reported more than 9.4 million new cases in 2015 and nearly 832,000 deaths in developed countries (2,3).
As a heterogeneous disease, CRC is associated with gene aberration, the microenvironment of tumor initiation, progression and metastasis (4). Over the last decade, many molecular biomarkers and signal pathways associated with the occurrence and progression of CRC have been reported, which has been involved in clinical therapy. To date, owing to the high incidence and mortality in the CRC disease, uncovering the causes and the molecular characterization of CRC to discover the molecular biomarkers for initial diagnosis, prevention and personalized therapy is quite urgent and demanded.
The recent high-throughput sequence techniques for the analyses of different gene expression and alternative splicing variants, like microarrays or the RNA-seq chip, are increasingly valued as promising techniques in medical oncology with great clinical applications, such as molecular diagnosis, prognosis prediction, drug targets discovery. Many gene expression profiling studies have been focused on CRC in the last decade (5), and hundreds of potential gene biomarkers have been obtained (6), which involved in different functional enrichments [including biological process (BP), cellular components (CC), molecular function (MF)], signal pathways and protein-protein interaction (PPI) networks.
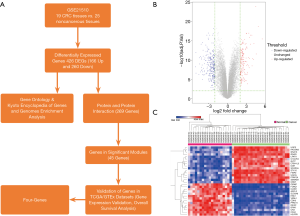
In this study, we have obtained the dataset (GSE21510) from National Center Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (NCBI-GEO) (website: https://www.ncbi.nlm.nih.gov/geo/), and we have chosen 25 CRC and 19 noncancerous tissue samples for differentially expressed genes (DEGs) analysis, gene ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, PPI networks complex construction, and hub genes exploration. What’s more, we have validated gene expression levels and overall survival (OS) analyses of these hub genes in related datasets [colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ)] built in the TCGA/GTEx database. Moreover, the filtered potential genes associated with CRC be recognized as biomarkers for diagnosis, prognosis and drug targets. Figure 1A shows the workflow of this study.
Methods
Data collection
The gene expression microarray data GSE21510 (7) were collected from the NCBI-GEO database (8,9). The GSE21510, taken the Affymetrix GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) as a reference, was submitted by Kaoru Mogushi et al., which featured 104 CRC patients. We choose 19 patients (CRC tissues, cancer group) and 25 patients (noncancerous tissues, normal group) to identify the genes and pathways.
Data preprocessing
After GSE21510 was obtained, probe identification numbers (IDs) were converted into gene symbols or ENTREZID. For multiple probes corresponding to the same one gene, their most significantly expressed value was treated as the gene expression value. Non-mRNA probes were discarded. Then, the gene expression values were normalized by using the Affy package, and RMA signal intensity was performed with log2 transformation and normalization (10,11).
Identification of DEGs
Linear models for microarray data (limma) is an R package applied to analyze gene expression matrix, especially when the linear models are constructed to assess the differentially expressed gene expression under the designed experiment condition (12). Limma package (http://bioconductor.org/packages/2.4/bioc/html/limma.html) built in R was applied to identify the DEGs between CRC tissues (cancer group) and noncancerous tissues (normal group). Significant DEGs were selected for further analyses with setting the |log2 fold change (FC)| ≥2 & the adjusted P value (adj.P.Val) <0.01 (13).
Function and signal pathway enrichment analysis
GO provides a controlled vocabulary of terms to elucidate a gene product’s characteristics via their annotation. GO terms reflect what is currently known about a gene in terms of BP, CC and MF (14,15). Moreover, KEGG (16) provides data resources of known biological pathways to annotate a gene or a set of genes/proteins with their respective KEGG pathways. In order to illustrate the function and signal pathway analysis of DEGs, GO and KEGG pathway enrichment analyses were carried out with using the clusterProfiler package and ReactomePA package (17,18) in R and P value <0.05 was considered significance.
PPI network and gene module analysis
Search Tool for the Retrieval of Interacting Genes (STRING) (19) is an open access database designed to evaluate the PPI messages of DEGs. STRING (version 10.5) covers 9.6 million proteins originated from 2,031 organisms. At first, we uploaded and mapped the list of DEGs to STRING website. Then PPIs of DEGs with a combined score >0.4 (medium confidence) and genes closely correlated with the other genes were chosen with the degree ≥10 (20). After that, PPI networks were constructed by using the Cytoscape software (21). The plug-in Molecular Complex Detection (MCODE) built in Cytoscape was applied to pick out the significant gene modules of the PPI networks. The parameters were set as follows: MCODE scores >3 and the count of nodes >4. Finally, we selected two significant gene modules (including 46 genes) from the PPI networks for further validation analyses.
Validation of four genes in TCGA/GTEx
To further screen for precise biomarkers, we have validated these 46 genes at gene expression level and OS time on web server GEPIA, which integrated COAD and READ datasets. The gene expression consistency and the survival analyses of these candidate genes were tested and evaluated in GEPIA. As for gene expression validation involved in 367 tumor samples (COAD: 275, READ: 92) and 667 normal samples (COAD: 349, READ: 318), the threshold with |log2 FC| ≥2 & P value <0.01 was considered statistically significant. For the OS analyses in integrated COAD & READ datasets, the 362 patients with available OS time data were sorted into low- and high-expression groups by the median transcripts per kilobase million (TPM), and significance was decided by the log-rank test with P<0.05.
Results
DEGs identification
Microarray data of 19 CRC tissues (cancer group) and 25 noncancerous tissues (normal group) were analyzed by the limma package built in R through the linear model and the contrast model to identify DEGs. A total of 426 DEGs were selected by the criteria of adjusted P value <0.01 & |log2 FC| ≥2, including 166 up-regulated genes and 260 down-regulated genes (Figure 1B, Table 1). The hierarchical cluster analysis was done to show the most 15 significantly up-regulated genes and 15 down-regulated genes in Figure 1C. The top ten genes with the most significant expression were CLDN1, CEMIP, UGP2, EPB41L3, CDH3, ENC1, PPM1H, GLTP, SLC6A6 and SEMA6A (Table 1).
Table 1
| DEGs | Gene symbol |
|---|---|
| Up-regulated | CLDN1, CEMIP, CDH3, ENC1, PPM1H, SLC6A6, ASCL2, VSNL1, FOXQ1, FABP6, PPAT, NUFIP1, NUDCD1, RAD54B, C2, HILPDA, MYC, CRNDE, NFE2L3, CKAP2, AJUBA, IRAK1BP1, CDK4, ATP11A, SLC7A5, EPHX4, LOC101060264, NEBL, PUS7, TRIB3, HOMER1, FAM60A, UTP14A, AXIN2, CSE1L, PPIL1, LYAR, DGAT2, SLC39A10, PAICS, CCDC113, TOP1MT, PROCR, MCM2, ZAK, ZC3HAV1L, HSP90AB1, TMPRSS3, UBE2C, LGR5, NUF2, RFC3, CDC25B, PRMT3, HELLS, DSCC1, PALD1, SLCO4A1, PLAU, ATAD2, BICD1, PARPBP, E2F7, TGFBI, PMAIP1, RPP40, AZGP1, DPEP1, KRT23, SPC25, MET, SP5, KIF20A, TRIP13, CDKN3, SLC12A2, PABPC1L, MMP1, ANLN, HS2ST1, CDK1, FAM92A1, CXCL3, SLC22A3, RNF43, ZNRF3, DUSP14, PSAT1, TPX2, ACSL4, AURKA, CKS2, MKI67, LDLRAD3, RAD51AP1, PHLDA1, SUPT16H, INHBA, SRPX2, MMP7, CXCL1, RASSF10, KIF4A, CXCL8, MMP3, TOP2A, BMP4, TESC, TTK, CTHRC1, HMMR, CDCA7, BUB1, DACH1, SLC38A5, CYP4X1, C2CD4A, GNG4, ELOVL5, CXCL2, FAM83D, CHI3L1, KLK10, TCFL5, LEF1, TACSTD2, DLGAP5, PRC1, ACSL6, NKD1, CADPS, NEK2, CENPK, MMP12, WT1, SLC35D3, CLDN2, PRR11, GINS1, COL11A1, CXCL10, CCNB1, CEP55, PTPRO, PLEKHB1, RGCC, CXCL11, SLCO1B3, GDF15, HS6ST2, RRM2, GAL, APCDD1, CXCL5, KRT6B, CKMT2, COL4A1, COL1A2, EDNRA, SPP1, TCN1, REG3A, AMIGO2, PPBP, WDR72, COL15A1 |
| Down-regulated | UGP2, EPB41L3, GLTP, SEMA6A, SCARA5, PPAP2A, UGCG, IL6R, RELL1, TP53INP2, GCNT2, SPPL2A, ETFDH, ADH1B, SGK1, MXI1, CNTN3, NR3C2, MMP28, ABCA8, ZZEF1, C2orf88, RHOU, TMEM220, SYTL4, KLF4, AGPAT9, SMIM14, KAT2B, ABCG2, METTL7A, TMCC3, FRMD3, SLC2A13, PARM1, SMPD1, ARRDC4, TSPAN7, ABI3BP, RUNDC3B, SLC30A4, SLC4A4, GRAMD3, ADCY9, SRI, HPGD, GUCA2A, MIER3, PLCE1, SLCO2A1, LIFR, PCSK5, PRKACB, FAM107B, TMEM100, WDR78, SCIN, FAM46A, MT1M, GUCA2B, SLC51B, PDE9A, PLCL2, SPIB, C1orf115, NR5A2, CDHR5, USP2, CA7, PADI2, CD177, BEST4, CPNE8, ENTPD5, HOXD1, PDE3A, PHLPP2, ANO5, CXCL12, SLC30A10, TEX11, KIAA1211, SEMA6D, SFRP1, TLCD2, TMEM56, RMDN2, MAOA, APPL2, ITM2C, MXD1, RHOF, NPY1R, SCNN1B, LDHD, PDK4, BHLHE41, PTPRH, BMP2, ADTRP, CCDC68, ARHGAP44, CHP2, ARHGAP42, PAG1, MT1E, PKIB, MEIS3P1, MT1HL1, MT1X, HDAC9, SSPN, SMPDL3A, ENDOD1, CLDN23, EDIL3, EPB41L4A, OSBPL1A, MT2A, HRCT1, MT1H, GBA3, CMAHP, SLC17A4, VLDLR, ITM2A, MALL, NAAA, GDPD3, AHCYL2, AQP8, TMEM72, CHST5, THRB, TBC1D9, SLC1A1, HSD17B2, SRPX, FGL2, HAGLR, CLU, CDHR2, KIF16B, VSIG2, CCL28, CA4, BEST2, MT1G, TNFSF10, MT1F, VILL, F2RL1, CES2, LAMA1, SLC41A2, RNF125, EGLN3, DHRS11, RETSAT, EMP1, OGN, CA2, CHRDL1, TMEM171, CWH43, CEACAM7, ST6GALNAC6, B3GALT5, LRRC19, ADH1C, FMO5, LGALS2, PTPRR, PRSS12, ARL14, LRRC66, PTGDR, SPON1, ZBTB7C, ANPEP, PIGR, EDN3, DHRS9, ZG16, HHLA2, PROM2, BCAS1, AKR1B10, CLCA4, LINC01133, BTNL8, MUC12, MALAT1, SLC16A9, TUBAL3, UGT2A3, HEPACAM2, NXPE4, CLIC6, TTC22, FAM134B, MS4A12, CA1, TRPM6, SI, CEACAM1, CYBRD1, IQGAP2, SLC16A14, SLC26A2, LYPD8, FOXP2, MYLK, HMGCS2, CFD, RASSF6, HSD11B2, GHR, AKAP7, DEFB1, DSC2, SULT1B1, PCK1, B3GNT7, NXPE1, MOGAT2, MEIS1, C4orf19, GGT6, IL1R2, MGP, SCNN1G, FHL1, LEPREL1, PLAC8, ENPP3, CLDN8, GCG, TSPAN1, ABCB1, NR1H4, MFAP5, FCGBP, VIP, RARRES1, SLC51A, SPINK5, CAPN13, INSL5, MEP1A, IGJ, GCNT3, ISX, MUC2, SYNPO2, AGR3, SFRP2, CLCA1, ITLN1, HSD3B2 |
Function and signal pathway enrichment analysis
To investigate functions and signal pathway enrichment of identified DEGs, we further analyzed DEGs using the clusterProfiler package and ReactomePA package in R with criteria of P<0.05. Figure 2A show the significant ten BP, CC and MF enrichment terms, respectively. Moreover, Figure 2B shows the top 30 significant GO terms.
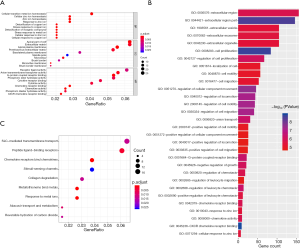
As shown in Table 2, in BP term, the up-regulated genes were mainly enriched in the nuclear division, mitotic nuclear division and organelle fission, while the down-regulated genes were focused on the detoxification of copper ion, stress response to copper ion and detoxification of inorganic compound. In CC term, the up-regulated genes were mainly enriched in the spindle pole, spindle, chromosome and centromeric region, while the down-regulated genes were focused on the microvillus membrane, apical part of cell and apical plasma membrane. In MF term, the up-regulated genes were mainly enriched in the CXCR chemokine receptor binding, chemokine activity and chemokine receptor binding, while the down-regulated genes were focused on the oxidoreductase activity, phosphoric diester hydrolase activity and oxidoreductase activity.
Table 2
| Terms | Category | Description | FDR | Count |
|---|---|---|---|---|
| Up-regulated genes | ||||
| GO: 0000280 | BP | Nuclear division | 4.27E-07 | 21 |
| GO: 0140014 | BP | Mitotic nuclear division | 5.16E-07 | 17 |
| GO: 0048285 | BP | Organelle fission | 9.75E-07 | 21 |
| GO: 0000922 | CC | Spindle pole | 0.000108 | 10 |
| GO: 0005819 | CC | Spindle | 0.000108 | 14 |
| GO: 0000775 | CC | Chromosome, centromeric region | 0.000574 | 10 |
| GO: 0045236 | MF | CXCR chemokine receptor binding | 2.53E-10 | 8 |
| GO: 0008009 | MF | Chemokine activity | 1.84E-06 | 8 |
| GO: 0042379 | MF | Chemokine receptor binding | 1.28E-05 | 8 |
| R-HSA-380108 | KEGG | Chemokine receptors bind chemokines | 7.09E-06 | 8 |
| R-HSA-1442490 | KEGG | Collagen degradation | 3.65E-05 | 8 |
| R-HSA-1474228 | KEGG | Degradation of the extracellular matrix | 0.001122 | 9 |
| Down-regulated genes | ||||
| GO: 0010273 | BP | Detoxification of copper ion | 5.97E-09 | 8 |
| GO: 1990169 | BP | Stress response to copper ion | 5.97E-09 | 8 |
| GO: 0061687 | BP | Detoxification of inorganic compound | 1.10E-08 | 8 |
| GO: 0031528 | CC | Microvillus membrane | 5.58E-05 | 6 |
| GO: 0045177 | CC | Apical part of cell | 5.58E-05 | 19 |
| GO: 0016324 | CC | Apical plasma membrane | 5.58E-05 | 17 |
| GO: 0016614 | MF | Oxidoreductase activity, acting on CH-OH group of donors | 0.00075 | 11 |
| GO: 0008081 | MF | Phosphoric diester hydrolase activity | 0.00075 | 9 |
| GO: 0016616 | MF | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 0.00075 | 10 |
| R-HSA-5661231 | KEGG | Metallothioneins bind metals | 1.59E-08 | 7 |
| R-HSA-5660526 | KEGG | Response to metal ions | 7.97E-08 | 7 |
| R-HSA-2672351 | KEGG | Stimuli-sensing channels | 0.000575 | 10 |
GO, gene ontology; FDR, false discovery rate; BP, biological process; CC, cellular components; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 2C shows the significant pathways in which the most signal pathways were enriched in the metallothioneins bind metals, response to metal ions and chemokine receptors bind chemokines (Figure 2C). The up-regulated genes were mainly enriched in the chemokine receptors bind chemokines, collagen degradation and degradation of the extracellular matrix (Table 2), and the down-regulated genes were enriched in the metallothioneins bind metals, response to metal ions and stimuli-sensing channels (Table 2).
Module screening from the PPI network
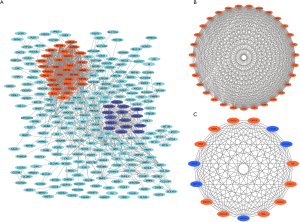
Firstly, 426 DEGs were uploaded into the STRING website and analyzed by Cytoscape software. A total of 269 DEGs with score >0.4 (medium confidence) were picked out for the construction of the PPI networks (Figure 3A). Then, two significant gene modules were clustered via MCODE APP built in Cytoscape. Module 1 was made up of 31 up-regulated genes/nodes and 427 edges (Figure 3B), while module 2 consisted of 15 genes/nodes (10 up-regulated and 5 down-regulated genes) and 105 edges (Figure 3C).
Validation of four genes in TCGA/GTEx
Genes from those two gene modules were chosen for validation in COAD and READ built in TCGA/GTEx datasets. We further found that only the four gene expressions of AURKA, BUB1, DLGAP5 and HMMR in COAD and READ datasets have consistency with that in GSE21510 (Figure 4A). In addition, their OS is significantly different between high expression and low expression (Figure 4B). GO and KEGG pathways show that these four candidate genes are significantly enriched in the regulation of mitotic cycle phase transition and oocyte meiosis (Figure 4C and Table 3).
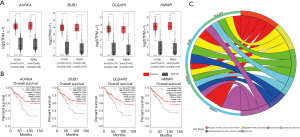
Table 3
| Terms | Category | Description | FDR | Gene | Count |
|---|---|---|---|---|---|
| GO: 1901990 | BP | Regulation of mitotic cell cycle phase transition | 0.00025 | AURKA, BUB1, DLGAP5, HMMR | 4 |
| GO: 0007088 | BP | Regulation of mitotic nuclear division | 0.00073 | AURKA, BUB1, DLGAP5 | 3 |
| GO: 0000280 | BP | Nuclear division | 0.001 | AURKA, BUB1, DLGAP5 | 3 |
| GO: 0030071 | BP | Regulation of mitotic metaphase/anaphase transition | 0.0031 | BUB1, DLGAP5 | 2 |
| GO: 0045840 | BP | Positive regulation of mitotic nuclear division | 0.0031 | AURKA, DLGAP5 | 2 |
| GO: 0032436 | BP | Positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 0.0058 | AURKA, DLGAP5 | 2 |
| GO: 1903047 | BP | Mitotic cell cycle process | 0.0058 | AURKA, BUB1, DLGAP5 | 3 |
| GO: 0051781 | BP | Positive regulation of cell division | 0.0066 | AURKA, DLGAP5 | 2 |
| GO: 0000819 | BP | Sister chromatid segregation | 0.0076 | BUB1, DLGAP5 | 2 |
| GO: 0140014 | BP | Mitotic nuclear division | 0.0082 | AURKA, DLGAP5 | 2 |
| GO: 0007093 | BP | Mitotic cell cycle checkpoint | 0.0089 | AURKA, BUB1 | 2 |
| GO: 0010389 | BP | Regulation of G2/M transition of mitotic cell cycle | 0.0089 | AURKA, HMMR | 2 |
| GO: 0140013 | BP | Meiotic nuclear division | 0.0089 | AURKA, BUB1 | 2 |
| GO: 1901991 | BP | Negative regulation of mitotic cell cycle phase transition | 0.0091 | AURKA, BUB1 | 2 |
| GO: 0051276 | BP | Chromosome organization | 0.0128 | AURKA, BUB1, DLGAP5 | 3 |
| GO: 0051301 | BP | Cell division | 0.0453 | AURKA, BUB1 | 2 |
| GO: 0031616 | CC | Spindle pole centrosome | 0.00041 | AURKA, DLGAP5 | 2 |
| GO: 0000780 | CC | Condensed nuclear chromosome, centromeric region | 0.00062 | AURKA, BUB1 | 2 |
| GO: 0005813 | CC | Centrosome | 0.0038 | AURKA, DLGAP5, HMMR | 3 |
| GO: 0043232 | CC | Intracellular non-membrane-bounded organelle | 0.0395 | AURKA, BUB1, DLGAP5, HMMR | 4 |
| hsa04114 | KEGG | Oocyte meiosis | 0.0028 | AURKA, BUB1 | 2 |
| hsa04914 | KEGG | Progesterone-mediated oocyte maturation | 0.0028 | AURKA, BUB1 | 2 |
GO, gene ontology; FDR, false discovery rate; BP, biological process; CC, cellular components; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
In this study, our purposes were to expound the potential genetic biomarkers and pathways through comparing the array datasets between cancer group (CRC tissues) and Normal group (noncancerous tissues), and 166 up-regulated and 260 down-regulated DEGs were identified. Then the function (GO) and signal pathway (KEGG) annotation analyses have been performed. Moreover, the PPI networks of DEGs were constructed and 269 DEGs/nodes were connected to 1,169 edges, and finally the most two significant modules were chosen from the PPIs, from which 46 central nodes/genes were selected to validate its gene expressions and OS time in TCGA/GTEx.
As we knew that mutations in the Wnt signaling pathway and inflammatory bowel disease are the two major causes of CRC (22). Through bioinformatics analyses, we have identified 46 central nodes/genes, among them, the first significant gene module (Figure 3B) consists of 31 genes, including TOP2A, CDK1, CCNB, MYC, AURKA, BUB1, etc., and the second significant gene module (Figure 3C) consists of 15 genes, including CXCL12, CCL28, CXCL2, GNG4, CXCL1, INSL5, etc. However, some of genes in these two significant gene modules, associated with CRC, have been researched and identified in the past years. Finally, we screened four genes (AURKA, BUB1, DLGAP5 and HMMR), which have been consistent with their gene expression level in tumor and normal patients of COAD & READ. Besides, we discarded the other 42 genes which didn’t meet the threshold we set.
Aurora A kinase (AUKRA) is encoded by the AURKA gene and a member of the serine/threonine kinases family (23,24). AURKA has been shown to interact with Wnt and Ras-MAPK signaling in CRC (25). What’s more, it was reported that AURKA has associated with CRC liver metastasis (CRLCM) (26,27). Budding uninhibited by benzimidazoles (BUB1) is also a member of the serine/threonine-protein kinase family (28). Over 90 percentages of all human solid tumors have a common feature that mutations occur in the spindle checkpoints (29). Jaffrey et al. have suggested that mutations in BUB1 can lead to chromosome instability in cancer cell lines (30). Disks large-associated protein 5 (DLGAP5) is a kinetochore protein that plays a role in stabilizing microtubules in chromosomes, controlling spindle dynamics, promoting interkinetochore tension and executing efficient kinetochore capture (31). However, Schneider MA’s team has predicted that AURKA and DLGAP5 could have a correlation with poor prognosis in non-small cell lung cancer patients (32). By regulating DLGAP5 gene expression, it could enhance the efficacy of epirubicin for invasive breast cancer (33). Hyaluronan-mediated motility receptor (HMMR), known as receptor for hyaluronan mediated motility (RHAMM), has a high-level gene expression and correlation with poor outcome in breast cancer (34). What’s more, HMMR may be associated with the risk of breast cancer patients who have BRCA1 mutation (35).
As shown in Figure 4B, you will be surprised to find that the low TPM group of these four genes have low percent survival than that in high TPM group, which are contrary to what we expected. But the survival curve of these four genes was statistically significant, and it is essential for us to seek the reasons why it occurs.
The above four genes were chosen via comprehensive bioinformatics analyses and mainly enriched in the regulation of mitotic cycle phase transition and oocyte meiosis pathway. However, further molecular and cellular experiments are required to verify the function of these four gene biomarkers in CRC.
Acknowledgments
Funding: This project was supported and funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. For human datasets mentioned in this study, please refer to the original article (PMID: 21270110). We just re-analyzed the open accessed datasets, and no ethical approval or informed consent was required by the local ethics committees. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Giardiello FM, Hamilton SR, Krush AJ, et al. Treatment of Colonic and Rectal Adenomas with Sulindac in Familial Adenomatous Polyposis. New Engl J Med 1993;328:1313-6. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Vos T, Allen C, Arora M, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1545-602. [Crossref] [PubMed]
- Aran V, Victorino AP, Thuler LC, et al. Colorectal Cancer: Epidemiology, Disease Mechanisms and Interventions to Reduce Onset and Mortality. Clin Colorectal Cancer 2016;15:195-203. [Crossref] [PubMed]
- Isella C, Terrasi A, Bellomo SE, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015;47:312-9. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Tsukamoto S, Ishikawa T, Iida S, et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res 2011;17:2444-50. [Crossref] [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009;37:D885-90. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Larriba Y, Rueda C, Fernández MA, et al. Microarray Data Normalization and Robust Detection of Rhythmic Features. In: Bolón-Canedo V, Alonso-Betanzos A. editors. Microarray Bioinformatics. New York, NY: Humana, 2019:207-25.
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3.
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003;19:368-75. [Crossref] [PubMed]
- Hale ML, Thapa I, Ghersi D. FunSet: an open-source software and web server for performing and displaying Gene Ontology enrichment analysis. BMC Bioinformatics 2019;20:359. [Crossref] [PubMed]
- Carbon S, Dietze H, Lewis S, et al. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 2017;45:D331-8. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;28:27-30.
- Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 2016;12:477-9. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Zhou Z, Cheng Y, Jiang Y, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci 2018;14:124-36. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Hu T, Li LF, Shen J, et al. Chronic inflammation and colorectal cancer: the role of vascular endothelial growth factor. Curr Pharm Des 2015;21:2960-7. [Crossref] [PubMed]
- Roskoski R Jr. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol Res 2019;139:471-88. [Crossref] [PubMed]
- Rozpędek W, Pytel D, Nowak-Zduńczyk A, et al. Breaking the DNA Damage Response via Serine/Threonine Kinase Inhibitors to Improve Cancer Treatment. Curr Med Chem 2019;26:1425-45. [Crossref] [PubMed]
- Jacobsen A, Bosch LJW, Martens-de Kemp SR, et al. Aurora kinase A (AURKA) interaction with Wnt and Ras-MAPK signalling pathways in colorectal cancer. Sci Rep 2018;8:7522. [Crossref] [PubMed]
- Goos JA, Coupe VM, Diosdado B, et al. Aurora kinase A (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. Br J Cancer 2013;109:2445-52. [Crossref] [PubMed]
- Goos JA, Coupe VM, van de Wiel MA, et al. A prognostic classifier for patients with colorectal cancer liver metastasis, based on AURKA, PTGS2 and MMP9. Oncotarget 2016;7:2123-34. [Crossref] [PubMed]
- Matsubara Y, Matsumoto T, Yoshiya K, et al. Budding uninhibited by benzimidazole-1 insufficiency prevents acute renal failure in severe sepsis by maintaining anticoagulant functions of vascular endothelial cells. Shock 2019;51:364-71. [Crossref] [PubMed]
- Williams BR, Amon A. Aneuploidy: cancer's fatal flaw? Cancer Res 2009;69:5289-91. [Crossref] [PubMed]
- Jaffrey RG, Pritchard SC, Clark C, et al. Genomic instability at the BUB1 locus in colorectal cancer, but not in non-small cell lung cancer. Cancer Res 2000;60:4349-52. [PubMed]
- Wilde A. "HURP on" we're off to the kinetochore! J Cell Biol 2006;173:829-31. [Crossref] [PubMed]
- Schneider MA, Christopoulos P, Muley T, et al. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol 2017;50:365-72. [Crossref] [PubMed]
- Zhang X, Pan Y, Fu H, et al. Nucleolar and Spindle Associated Protein 1 (NUSAP1) Inhibits Cell Proliferation and Enhances Susceptibility to Epirubicin In Invasive Breast Cancer Cells by Regulating Cyclin D Kinase (CDK1) and DLGAP5 Expression. Med Sci Monit 2018;24:8553-64. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A 2004;101:9309-14. [Crossref] [PubMed]
- Blanco I, Kuchenbaecker K, Cuadras D, et al. Assessing associations between the AURKA-HMMR-TPX2-TUBG1 functional module and breast cancer risk in BRCA1/2 mutation carriers. PLoS One 2015;10:e0120020. [Crossref] [PubMed]


