
Over-expression of HDAC8 down-regulate CDKN2A is associated with worse prognosis of esophageal squamous cell carcinoma
Introduction
Esophageal cancer (EC) one of the most complex and progressive cancers that are the 8th most common and 6th most deadly cancer throughout the world (1). Within two histological subtypes of EC, which is esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), the EAC is more common in the western world, while ESCC is more predominant in central and southeastern Asia, mostly in China and Japan (2). Only in China, 477,900 newly detected EC cases, and 375,000 mortalities from EC were documented to occur in 2015 (3). Notwithstanding current encroachment in combined treatment schemes, including surgery, chemotherapy, and radiotherapy, the outcome for EC patient’s still substandard, mainly for advanced-stage patients (4). As well as 5-year overall survival (OS) rate yet at less than 20% (5). The hastily rising prevalence, challenging management, poor prognosis of ESCC in emphasizing the necessity for effectual potential biomarkers for prompt diagnosis, prognosis evaluation, and narrative therapeutic targets. The etiology and pathogenesis of ESCC encompass complex interfaces between epigenetic, genetic, and environmental causes (6-8). DNA methylation and histone deacetylation are communal forms of epigenetic modification, that have a critical role in various carcinomas, including breast cancer, lung cancer, and gastric cancers (9-11). Targeted gene modification and transcription occur due to aberrant changes in the promoter region of tumor suppressor genes. Furthermore, DNA methylation and deacetylation has performed as an independent promising biomarker for early detection and diagnosis of cancers (12).
One of the tumor suppressor proteins that are inactivated in carcinoma is the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene, which located within the frequently deleted chromosome 9p21 (13). It is responsible for inhibiting various cyclin-dependent kinases and plays an important role in cell cycle regulation by decelerating cell cycle progression at the G1/S phase (14,15). Deacetylation of histone and hyper-methylation of the CDKN2A gene promoter segment causes its inactivation, ultimately develop several kinds of malignancies such as lung carcinoma, head and neck carcinoma, hepatocellular carcinoma, breast carcinoma, esophageal carcinoma, and so on (16-20). Epigenetic alterations such as DNA methylation or histone modifications and so on are advocated to synchronize gene expression without affecting the base sequence (21-23). Silenced CDKN2A reactivation or the epigenetic repression capacity inhibition could be a coherent policy for the prevention or treatment of many carcinomas. However, this phenomenon of epigenetic silencing of CDKN2A is due to a histone modification and renewal of p16 transcriptional stimulation can be attained by a group of intracellular regulatory genes (24-26). Epigenetic modification comprising histone acetylation and deacetylation regulated by either histone deacetylase’s (HDACs) or histone acetyltransferase (HAT) (27). Within 18 HDACs, HDAC8 is one of the most culprit agents that overexpressed in carcinoma, causes epigenetic alteration and has an impact on cancer cell cycle mostly in tumor suppressor in CDKN2A and ultimately, cancer progression rapidly.
Many pieces of research held about epigenetic modification and effect on tumor suppressor gene and their effect on survival status. But this is the first time we do study about this on ESCC. Our study aims to determine the expression status of HDAC8 affect expression status of CDKN2A and their effect on the prognostic value and disease progression.
Methods
Patients and tumor samples
One hundred and thirty-five cases of ESCC were randomly and retrospectively selected from the files of the department of pathology of Qilu Hospital of Shandong University and tissue samples obtained from patients at subtotal esophagectomy and esophagogastric anastomosis with regional lymph node dissection at the year of 2010 and 2011. For our research, we designed inclusion criteria such as, didn’t receive any chemotherapy or radiotherapy or immunotherapy preoperatively, all the tumor stages, any lymph node status; and exclusion criteria such as younger age group (less than 25 years), and already receive any single or combined therapy before surgery. From our Qilu hospital database, we accumulated all the baseline, demographic and investigational data of all patients. The patients who didn’t meet the inclusion criteria and also lost in follow up were excluded from the final study group and 110 patients were selected for our study. For tumor staging, we accustomed “American Joint Committee on Cancer (AJCC) Staging Manual, 7th edition, 2010”. Our study design, tissue sample, and data collection were accomplished according to our institutional protocols, which approved by the ethics board of Qilu Hospital of Shandong University. All the patients who include in our study give written informed consent.
Follow-up of study population
The included patients were followed-up routinely either till their expiry or at least 5 years from their surgery date. At the time of follow-up thorough physical examination and routine imaging, investigation was performed every three months up to 1st two years after done surgery and every 6 months up to the next 3 to 5 years. Depend on the investigation and examination report; routine radiological examination and esophagoscopy were done. The other data collected from the database for in-patients or from the outpatient department tumor registry of our hospital.
Immunohistochemical staining
All the fresh tissue samples were collected from the patients during operation in the year of 2010 and2011. The paraffin-embedded fresh specimens were fixed by 10% formalin that later we collected from the pathology department of the Qilu Hospital of Shandong University. We collected two sets of 110 tissue samples for our two biomarker HDAC8 and CDKN2A. The collected tissues were cut section 4-mm by serial from paraffin embed and then tissue samples retrieved by 10 mM citrate buffer. After retrieved samples were deparaffinized by Xylene and then did rehydration. Tissue sections were incubated in 3% H2O2 by methanol at least 20 minutes at room temperature. After that one set of tissue sample incubated again with primary anti-CDKN2A monoclonal antibody ab108349 (1:150, Abcam, Cambridge, MA, USA) and another set of tissue sample incubated by primary anti-HDAC8 polyclonal antibody ab217702 (1:150, Abcam, Cambridge, MA, USA); overnight in the high humid chamber at 4 °C. The next day morning the slides were again incubated at 37 °C for 30 minutes by biotinylated secondary antibodies and streptavidin-peroxidase complex. At the end of this immunohistochemistry method, slides were counterstained with 3,3’-diaminobenzidine solution with hematoxylin. After that fix the slides with a coverslip using natural balsam and let them dry. We used PBS instead of the primary antibody for the incubation of tissue samples for guided to negative controls.
Method of interpretation of immunohistochemistry and study endpoint
After dry off the immunostained slides, two skilled pathologists and we examined that under a light microscope. And independently scored by two investigators and the scores that were contradictory, sort out by them firmly. The staining intensity calculated by Immunoreactivity Score (IRS) system where the scoring parameter divided as: 0= no staining, 1= weak staining, 2= moderate staining, and 3= strong staining. After scorning the outcome calculated by multiplication of staining intensity by % of positive cells. We scored the positive cell to count positive cell, and positive scored as: 0=0–10% positive cells, 1=10–25% positive cells, 2=26–50% positive cells, 3=51–75% positive cells, and 4=76–100% positive cells. For calculating the final result, we did a summation of staining intensity and the % of the positive cells. That was further divided into four group; negative (−), weak (+), moderate (++), and strong (+++) respectively (0 to 1), (2 to 3), (4 to 5), and (6 to 7) (Figures 1,2). After calculation, base on expression status we divide our study group into “a weak or non-expression (− or +)” group and “high expression (++ or +++) group.
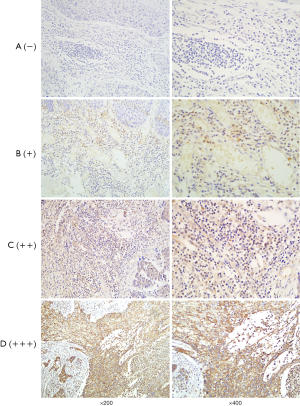
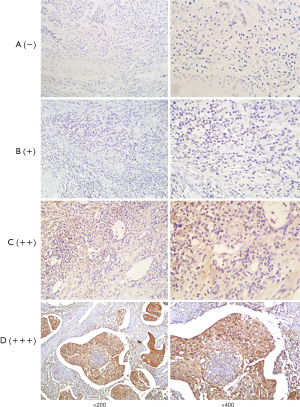
Our primary endpoint of the study was OS that is stated as the time from the date of surgery to death or the last follow-up date. And the secondary endpoint was progression-free survival (PFS) that denotes the local progression of the disease from the date of surgery to local or distant progression of the disease.
Statistical analysis
For statistical analysis, we categorized our study group into two groups; weak or non-expression group and highly expressed group. Associations of HDAC8 and CDKN2A with clinical parameters were analyzed by the Chi-square test or Fisher’s exact test. OS and PFS were estimated at 5 years after surgery with the Kaplan-Meier method and compared with HDAC8 and CDKN2A expression. For the determined independent prognostic factors, we use multivariate Cox proportional hazards regression models. Estimates at 5 years and 95% CI were reported on account of sample size, the timing of events, and available follow-up time. Spearman rank test was done due to determine the correlations between two prognostic markers. Our statistical analyses were performed operating SPSS statistics version 23.0 (SPSS Inc, Chicago, IL). All the tests were 2-sided and P values were significant when P<0.05.
Results
Staining assortment
We examined HDAC8 and CDKN2A protein expression of ESCC by immunohistochemical staining (IHC) technique of tissue microarray where we include 110-FFPE tissue samples for each protein (Figures 1,2). After reviewing the result of immunohistochemistry, it showed out of 110 patients HDAC8 overexpressed in 85.45% (94 patients) and for CDKN2A 80% (88 patients) patients are weak or non-expressed.
Demographic features of ESCC patients
A total of 110 patients from 135 patients, were met the criteria of our study design. The median age of our study population was 65 years, ranging from 25 to 86 years. Out of the 40 were female and 70 were male patients. Median follow-up duration of patients was 71 months, ranging from 1 to 122 months. We include all four tumor stages, where 20 cases from stage T1, 43 from stage T2, 35 from stage T3 and 20 cases from stage T4 were documented. Furthermore, lymph node status staged as no positive lymph node (N0) includes 44 cases, stage N1 24 cases, stage N2 includes 29 cases and stage N3 includes 13 cases. The degree of differentiation of tumor also analyzed in our study, out of all patients, well-differentiated tumor 32 cases, moderately differentiated tumor 31 cases and poorly differentiated tumor 47 cases reported (Table 1).
Table 1
| Biomarkers | Number (%) of patients with weak or no expression | Number (%) of patients with high expression |
|---|---|---|
| CDKN2A (p16) | 88 (80%) | 22 (20%) |
| HDAC8 | 16 (14.54%) | 94 (85.45%) |
Association between HDAC8 and CDKN2A with baseline characteristics
There were significant correlations between HDAC8 and CDKN2A protein expression with demographic data of our research population that analyzed by bilateral χ2 test. Most of ESCC patients of our study showed HDAC8 over-expression, from them, stage T1 11 patients (10% within total 110 patients), T2 38 patients (34.5%), T3 28 patients (25.5%) and T4 17 (15.5%). As well as lymph node status; stage N0 35 patients (31.8% within total 110 patients), stage N1 20 patients (18.2%), stage N2 26 patients (23.6%), stage N3 13 patients (11.8%). For most cases CDKN2A showed weak or no expression, tumor stage T1 includes 6 patients (5.5% within all 110 patents), stage T2 36 patients (32.7%), stage T3 27 patients (24.5%), and stage T4 19 patients (17.3%). As well as, lymph node station N0 includes 31 patients (28.2% within all 110 cases), stage N1 17 patients (15.5%), stage N2 27 patients (24.5%), and stage N3 includes 13 patients (11.8%). Here, both HDAC8 and CDKN2A are statistically significant for the lymph nodal stage but HDAC8 wasn’t significant for tumor stage whereas CDKN2A was significant. All the demographic data of 110 ESCC patients were summarized in Table 2.
Table 2
| Variables | All cases | HDAC8 expression (% within total cases) | CDKN2A expression (% within total cases) | |||||
|---|---|---|---|---|---|---|---|---|
| Weak or no | High | P* | Weak or no | High | P* | |||
| Age (years) | ||||||||
| <65 | 49 | 7 (6.4) | 42 (38.2) | 0.5 | 37 (33.6) | 12 (10.9) | 0.3 | |
| >65 | 61 | 9 (8.2) | 52 (47.3) | 51 (46.4) | 10 (9.1) | |||
| Gender | ||||||||
| Male | 40 | 4 (3.6) | 36 (32.7) | 0.4 | 36 (32.7) | 4 (3.6) | 0.05 | |
| Female | 70 | 12 (10.9) | 58 (52.7) | 52 (47.3) | 18 (16.4) | |||
| Smoking | ||||||||
| Smoker | 64 | 12 (10.9) | 52 (47.3) | 0.1 | 50 (45.5) | 14 (12.7) | 0.6 | |
| Non-smoker | 46 | 4 (3.6) | 42 (38.2) | 38 (34.5) | 8 (7.3) | |||
| Drinking habit | ||||||||
| Alcoholic | 56 | 11 (10.0) | 45 (40.9) | 0.1 | 41 (37.3) | 15 (13.6) | 0.09 | |
| Non-alcoholic | 54 | 5 (4.5) | 49 (44.5) | 47 (42.7) | 7 (6.4) | |||
| Tumor stages | ||||||||
| T1 | 12 | 1 (0.9) | 11 (10.0) | 6 (5.5) | 6 (5.5) | |||
| T2 | 43 | 5 (4.5) | 38 (34.5) | 0.6 | 36 (32.7) | 7 (6.4) | 0.01 | |
| T3 | 35 | 7 (6.4) | 28 (25.5) | 27 (24.5) | 8 (7.3) | |||
| T4 | 20 | 3 (2.7) | 17 (15.5) | 19 (17.3) | 1 (0.9) | |||
| Nodal station | ||||||||
| N0 | 44 | 9 (8.2) | 35 (31.8) | 31 (28.2) | 13 (11.8) | |||
| N1 | 24 | 4 (3.6) | 20 (18.2) | 0.05 | 17 (15.5) | 7 (6.4) | 0.003 | |
| N2 | 29 | 3 (2.7) | 26 (23.6) | 27 (24.5) | 2 (1.8) | |||
| N3 | 13 | 0 (0.0) | 13 (11.8) | 13 (11.8) | 0 (0.0) | |||
| Differentiation | ||||||||
| Poor | 47 | 8 (7.3) | 39 (35.5) | 36 (32.7) | 11 (10.0) | |||
| Moderate | 31 | 4 (3.6) | 27 (24.5) | 0.8 | 26 (23.6) | 5 (4.5) | 0.7 | |
| Well | 32 | 4 (3.6) | 28 (25.5) | 26 (23.6) | 6 (5.5) | |||
| Survival | ||||||||
| Alive | 50 | 14 (12.7) | 36 (32.7) | 0.001 | 29 (26.4) | 21 (19.1) | 0.001 | |
| Dead | 60 | 2 (1.8) | 58 (52.7) | 59 (53.6) | 1 (0.9) | |||
| Progression of disease | ||||||||
| Progression | 69 | 2 (1.8) | 67 (60.9) | 68 (61.8) | 1 (0.9) | |||
| No progression | 41 | 14 (12.7) | 27 (24.5) | 0.001 | 20 (18.2) | 21 (19.1) | 0.001 | |
*, Chi-square test. FFPE, formalin-fixed paraffin-embedded.
Over-expression of HDAC8 and loss of expression of CDKN2A correlated with worse outcomes in ESCC patients and correlation between these two biomarkers
We observe HDAC8 highly regulated and CDKN2A lost its expression capacity in ESCC patients and it has a worse effect on patients’ survival rate and disease progression and recurrence. From our total research population, 60 patients died during the follow-up period. Out of 60 died patients 58 patients have HDAC8 over-expression which was 52.7% of total 110 patients and 59 patients lost their CDKN2A expression capability, which was 53.6% of total 110 patients, and both were statistically significant, P=0.001. The median survival month for our study population was 42 months, ranging from 6 to 78 months. The prognostic importance and correlation between HDAC8 and CDKN2A with ESCC were analyzed by univariate analysis with Kaplan-Meier estimates using the log-rank test. Therefore, our analysis showed that high expression of HDAC8 significantly lowers the 5-year OS (P=0.003) and 5-year PFS (P=0.005) (Table 3, Figure 3A,B). As well as, CDKN2A showed low expression that also significantly lower the 5-year OS (P=0.001) and 5-year PFS (P=0.001) (Table 3, Figure 3C,D). The multivariate Cox-regression method used in our analysis to determine the independent prognostic marker and its further correlation. Regression analysis showed that 5-year OS and PFS of HDAC8 statistically significant, HR 1.173 with 95% CI: 0.215–6.416 and P=0.003 and HR 1.217 with 95% CI: 0.224–6.600 and P=0.005 respectively. Also another indicator, CDKN2A that is statistically significant, showed 5-year OS and PFS respectively as HR 0.65 with 95% CI: 0.680–1.468 and P=0.002 and HR 0.53 with 95% CI: 0.524–0.551 and P=0.001. The Spearman rank test was done due to clear about the correlation between HDAC8 and CDKN2A and there was a negative and inverse correlation between them. Table 4 showed the correlation coefficient was (−0.696). The univariate analysis displayed that among analyzed demographic parameters, traditional prognostic marker including tumor stage (P=0.001), lymph node (P=0.001), and drinking habit (P=0.01) were statistically significant in association with OS and tumor stage (P=0.03) and lymph node status (P=0.01) in association with PFS (Table 3). As well as we further investigate our indicators by multivariate cox-regression analysis, statistical significance for tumor stage HR 0.407 with 95% CI: 0.220–0.754, P=0.004 and HR 0.504 with 95% CI: 0.288–0.883 in association with OS and PFS respectively (Table 3).
Table 3
| Variables | OS | PFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | ||||||
| P | HR (95% CI) | P | P | HR (95% CI) | P | ||||
| Age (<65 vs. >65) | 0.6 | 1.014 (0.588–1.749) | 0.9 | 0.02 | 1.148 (0.691–1.907) | 0.5 | |||
| Gender (male vs. female) | 0.9 | 0.87 (0.485–1.559) | 0.6 | 0.08 | 0.825 (0.472–1.441) | 0.4 | |||
| Smoking (smoker vs. non-smoker) | 0.5 | 1.086 (0.577–2.046) | 0.7 | 0.1 | 1.246 (0.683–2.273) | 0.4 | |||
| Drinking habit (alcoholic vs. non-alcoholic) | 0.01* | 1.030 (0.554–1.915) | 0.9 | 0.5 | 0.842 (0.468–1.514) | 0.5 | |||
| Tumor stage (T3 & T4 vs. T1 & T2) | 0.001* | 0.407 (0.220–0.754) | 0.004* | 0.03* | 0.504 (0.288–0.883) | 0.01* | |||
| Nodal stage (N2 & N3 vs. N0 & N1) | 0.001* | 0.814 (0.472–1.402) | 0.4 | 0.01* | 0.679 (0.407–1.133) | 0.1 | |||
| Differentiation (well vs. moderate & poor) | 0.2 | 0.897 (0.644–1.249) | 0.5 | 0.1 | 0.889 (0.654–01.210) | 0.4 | |||
| HDAC8 (high expression vs. weak & no expression | 0.003* | 1.173 (0.215–6.416) | 0.003* | 0.005* | 1.217 (0.224–6.600) | 0.005* | |||
| CDKN2A (high expression vs. weak & no expression) | 0.001* | 0.65 (0.680–1.468) | 0.002* | 0.001* | 0.53 (0.524–0.551) | 0.001* | |||
*, statistically significant P value. OS, overall survival; PFS, progression free survival; CI, confidence interval.
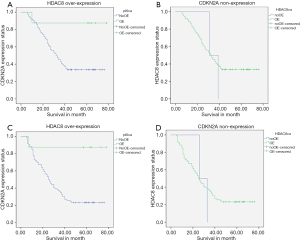
Table 4
| Prognostic factor | Spearman’s Rho |
|---|---|
| CDKN2A (p16) | 1 |
| HDAC8 | −0.696 |
| P value | 0.001 |
Discussion
Within our best knowledge, this is the first study that demonstrated highly regulated HDAC8 alter the normal cell cycle by down-regulation of CDKN2A protein and that causes cancer progression, metastases, tumorigenesis, and ultimately poor OS and worse prognosis. Here we will discuss more the mechanism with documents and according to research results.
Carcinoma conciliations an assortment of composite genetic and epigenetic events, which arise by several processes (28). The cell cycle is regulated by various proteins that precisely maintain the cell cycle checkpoints (26,29). These proteins may be deregulated in cancer as an effect of down regulation of various tumor suppressor genes (29,30). Mainly the event of loss of the tumor suppressor genes and their programmed proteins occurs through deletion, inactivating mutations, epigenetic silencing or post-translational modification that causes tumorigenesis. The advancement of the cell cycle from GI to mitosis is controlled by numerous cyclin protein and their catalytic subunits stated to as Cyclin-dependent kinases (CDKs) (31). CDKN2A is one of the important family members of this kinase family.
Epigenetic modifications alter the inherent status of gene expression and its mechanism control all biological procedure from the conception to death, by instituting epigenetic marker. Epigenetic mechanisms such as DNA methylation and histone modification have been shown to affect the transcription of key genes involved in the regulation of cellular growth, differentiation, apoptosis, and transformation, and tumor progression.
During the last few decades, CDKN2A has been one of the most comprehensively studied tumor suppressor genes. It plays a crucial role in cell cycle progression, and apoptosis (32,33). It acts as an inhibitor of the CDK4 (cyclin-dependent kinase 4) family of the cell cycle regulatory kinases (26,34). The histone modifications have some role to play in transcriptional regulation and so each has the impending to be oncogenic while there is loss of expression of a tumor suppressor gene (35). Histone deacetylase (HDACs) plays a vital role as a transcriptional repressor that alters in various cancers (36). Methylation and deacetylation associated gene silencing have been determined in various genes, CDKN2A one out of them and his protein has a direct link between DNA methylation and histone modification (37,38). Histones replicated and reoriented with accurate post-translational modification and variations in the metabolic programs and status of different cell types determine the level of chromatin compaction and exposure of DNA to replication errors, which regulates the cell genomic integrity and sensitivity to DNA damage (39).
Epigenetic regulators are commonly deregulated in cancers (40). Within 18 HDACs from HDACs family, HDAC8 overexpressed in numerous cancers, such as gastric cancer, colorectal, breast, prostate, thymic cancer, hepatocellular cancer, lung cancer, pancreas cancer and so on (41-45). In breast and prostate cancer HDACs overexpressed causes increase histone modification and transcriptional repression of suppressor genes such as CDKN2A (46). HDAC8, which is a member of class 1 HDAC showed over-expression in immunohistochemistry. Examination of the association between HCC progression and HDAC8 expression revealed a positive correlation of HDAC8 expression with tumor size, invasion and progression, ultimately poor OS and HDAC8 also repress the activity of tumor suppressor gene, by this alteration of the cancer cell cycle (47,48). In colorectal cancer (CRC), HDACs are frequently overexpressed and targets of anticancer therapy. HDAC8 highly expressed that deregulates tumor suppressor gene CDKN2A and causes the progression of the disease (49).
In our research, HDAC8 protein was overexpressed in most of the patients mostly in advance stage and deceased patients; whereas CDKN2A lost its expression capacity. After analysis, it comes in conclusion that the expression of HDAC8 protein and CDKN2A has inverse correlation and HDAC8 overexpressed and CDKN2A down-regulate indicates worst prognosis as well as disease progression and metastasis.
Past decades various studies held on ESCC and epigenetic alteration and tumor suppressor gene repression and observe their effect on disease outcome. Simão Tde et al. reported that lower expression of p16 (CDKN2A) correlates with higher DNA methylase expression (50). Zhao et al. demonstrated the implication of genetic and epigenetic alterations (HDAC8) of CDKN2A in many cancers such as esophageal, gastric, skin cancer, lung, pancreatic, head and neck, colorectal cancer, ovarian, prostate, renal cancer (51). So, reactivation of silenced CDKN2A or the inhibition of epigenetic repression of the gene could be a rational strategy for the prevention or treatment of ESCC.
Conclusions
Our results revealed a complex pattern of interactions between over-expression of HDAC8 and low or no expression of CDKN2A and its correlation with the outcome of ESCC, which indicates poor prognosis and more progression of ESCC. Also, over-expression of HDAC8 and weak expression of CDKN2A positively correlate with positive lymph node and advance tumor stage. Moreover, HDAC8 and CDKN2A act as promising pre and postoperative biomarkers and help to determine the further treatment plan.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.32). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Qilu Hospital of Shandong University. Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Wong MCS, Hamilton W, Whiteman DC, et al. Global Incidence and mortality of oesophageal cancer and their correlation with socioeconomic indicators temporal patterns and trends in 41 countries. Sci Rep 2018;8:4522. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Kauppila JH, Mattsson F, Brusselaers N, et al. Prognosis of oesophageal adenocarcinoma and squamous cell carcinoma following surgery and no surgery in a nationwide Swedish cohort study. BMJ Open 2018;8:e021495. [Crossref] [PubMed]
- Xu C, Wang Y, Yang H, et al. Association Between Cancer Incidence and Mortality in Web-Based Data in China: Infodemiology Study. J Med Internet Res 2019;21:e10677. [Crossref] [PubMed]
- Yousef M, Sharifi-Esfahani M, Pourgholam-Amiji N, et al. Esophageal cancer in the world: incidence, mortality and risk factors. Bio Res Ther 2018;5:2504-17. [Crossref]
- Yang X, Chen X, Zhuang M, et al. Smoking and alcohol drinking in relation to the risk of esophageal squamous cell carcinoma: A population-based case-control study in China. Sci Rep 2017;7:17249. [Crossref] [PubMed]
- Yang CX, Wang HY, Wang ZM, et al. Risk Factors for Esophageal Cancer: a Case-control Study in Southwestern China. Asian Pac J Cancer Prev 2005;6:48-53. [PubMed]
- Wu L, Shen Y, Peng X, et al. Aberrant promoter methylation of cancer-related genes in human breast cancer. Oncol Lett 2016;12:5145-55. [Crossref] [PubMed]
- Kajiura K, Masuda K, Naruto T, et al. Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget 2017;8:2890-905. [Crossref] [PubMed]
- Wang N, Sui F, Ma J, et al. Site-specific hypermethylation of RUNX3 predicts poor prognosis in gastric cancer. Arch Med Res 2016;47:285-92. [Crossref] [PubMed]
- Arantes LM, de Carvalho AC, Melendez ME, et al. Methylation as a biomarker for head and neck cancer. Oral Oncol 2014;50:587-92. [Crossref] [PubMed]
- Semczuk A, Boltze C, Marzec B, et al. p16 INK4A Alterations are accompanied by aberrant protein immunostaining in endometrial carcinomas. J Cancer Res Clin Oncol 2003;129:589-96. [Crossref] [PubMed]
- Park MT, Lee SJ. Cell Cycle and Cancer. J Biochem Mol Biol 2003;36:60-5. [PubMed]
- VanArsdale T, Boshoff C, Arndt KT, et al. Abraham. Molecular Pathways: Targeting the Cyclin D-CDK4/6 Axis for Cancer Treatment. Clin Cancer Res 2015;21:2905-10. [Crossref] [PubMed]
- Tam KW, Zhang W, Soh J, et al. CDKN2A/p16 inactivation mechanisms and their relationship to smoke exposure and molecular features in non-small-cell lung cancer. J Thorac Oncol 2013;8:1378-88. [Crossref] [PubMed]
- Pierini S, Jordanov SH, Mitkova AV, et al. Promoter hypermethylation of CDKN2A, MGMT, MLH1, and DAPK genes in laryngeal squamous cell carcinoma and their associations with clinical profiles of the patients. Head Neck 2014;36:1103-8. [Crossref] [PubMed]
- Csepregi A, Ebert MP, Rocken C, et al. Promoter methylation of CDKN2A and lack of p16 expression characterize patients with hepatocellular carcinoma. BMC Cancer 2010;10:317. [Crossref] [PubMed]
- Sinha S, Chunder N, Mukherjee N, et al. Frequent deletion and methylation in SH3GL2 and CDKN2A loci are associated with early- and late-onset breast carcinoma. Ann Surg Oncol 2008;15:1070-80. [Crossref] [PubMed]
- Xu R, Wang F, Wu L, et al. A systematic review of hypermethylation of p16 gene in esophageal cancer. Cancer Biomark 2013;13:215-26. [Crossref] [PubMed]
- Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci 2013;14:4655-69. [Crossref] [PubMed]
- Rodríguez-Paredes M, Esteller M. The Fundamental Role of Epigenetic Regulation in Normal and Disturbed Cell Growth, Differentiation, and Stemness. In: Lübbert M, Jones P. (eds). Springer: Epigenetic Therapy of Cancer, 2014.
- Sarkar D, Leung EY, Baguley BC, et al. Epigenetic regulation in human melanoma: past and future. Epigenetics 2015;10:103-21. [Crossref] [PubMed]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011;17:330-9. [Crossref] [PubMed]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002;298:1039-43. [Crossref] [PubMed]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell 2007;130:223-33. [Crossref] [PubMed]
- de Ruijter AJ, van Gennip AH, Caron HN, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003;370:737-49. [Crossref] [PubMed]
- Tallen G, Riabowol K. Keep-ING balance: tumor suppression by epigenetic regulation. FEBS Lett 2014;588:2728-2742. [Crossref] [PubMed]
- Williams GH, Stoeber K. The cell cycle and cancer. J Pathol 2012;226:352-64. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2002;144:646-74. [Crossref] [PubMed]
- Nabel EG. CDKs and CKIs: molecular targets for tissue remodeling. Nat Rev Drug Discov 2014;1:587-98. [Crossref] [PubMed]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell 2006;127:265-75. [Crossref] [PubMed]
- Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res 2005;576:22-38. [Crossref] [PubMed]
- Wang X, Pan L, Feng Y, et al. P300 plays a role in p16 (INK4a) expression and cell cycle arrest. Oncogene 2008;27:1894-904. [Crossref] [PubMed]
- Zhang K, Dent SY. Histone modifying enzymes and cancer: going beyond histones. J Cell Biochem 2005;96:1137-48. [Crossref] [PubMed]
- Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer 2005;41:2381-402. [Crossref] [PubMed]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143-53. [Crossref] [PubMed]
- Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457-463. [Crossref] [PubMed]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 2005;15:490-5. [Crossref] [PubMed]
- Nair N, Shoaib M, Sorenson CS. Chromatin Dynamics in Genome Stability: Roles in Suppressing Endogenous DNA Dam-age and Facilitating DNA Repair. Int J Mol Sci 2017;18:1-21. [Crossref] [PubMed]
- Weichert W, Röske A, Gekeler V, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol 2008;9:139-48. [Crossref] [PubMed]
- Weichert W, Roeske A, Niesporek S, et al. Class I histone de- acetylase expression has independent prognostic impact in human colorectal cancer—specific role of class I HDACs in vitro and in vivo. Clin Cancer Res 2008;14:1669-77. [Crossref] [PubMed]
- Weichert W, Röske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer 2008;98:604-10. [Crossref] [PubMed]
- Krusche CA, Wülfing P, Kersting C, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat 2005;90:15-23. [Crossref] [PubMed]
- Nakagawa M, Oda Y, Eguchi T, et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep 2007;18:769-74. [PubMed]
- Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis 2010;31:27-36. [Crossref] [PubMed]
- Wang LT, Chiou SS, Chai CY, et al. Aryl hydrocarbon receptor regulates histone deacetylase 8 expression to repress tumor suppressive activity in hepatocellular carcinoma. Oncotarget 2017;8:7489-501. [PubMed]
- Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK–cyclin complexes by INK4 inhibitors. Genes Dev 2000;14:3115-25. [Crossref] [PubMed]
- Wilson AJ, Byun DS, Popova N, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem 2006;281:13548-58. [Crossref] [PubMed]
- Simão Tde A, Simões GL, Ribeiro FS, et al. Lower expression of p14ARF and p16INK4a correlates with higher DNMT3B expression in human esophageal squamous cell carcinomas. Hum Exp Toxicol 2006;25:515-22. [Crossref] [PubMed]
- Zhao R, Choi BY, Lee MH, et al. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine 2016;8:30-9. [Crossref] [PubMed]


