
NLRC5, a valuable marker for the diagnosis and prognostic assessment of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is a highly malignant tumor with a poor prognosis (1,2). Although there exists a comprehensive diagnostic strategy that has been widely applied in the clinical setting, the early diagnosis of HCC is still a challenge (3,4). Also, there is currently no marker for the prognostic assessment of HCC. Thus, there is an urgent need to find a reliable marker for the early diagnosis and prognostic assessment for HCC.
Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are intracellular sensors of molecules associated with pathogens, which can trigger a hypersensitive response once a pathogen is detected (5,6). As a component of inflammasome, NLRs are involved in dysregulated inflammation and the initiation and progression of cancers (7,8).
As a member of the NLR family, NLR family CARD domain containing 5 (NLRC5) can effectively regulate the body’s immune response by negatively regulating the nuclear factor-kappa B subunit (NF-κB) and interferon (IFN) pathways (9). Additionally, enhanced NLRC5 expression has been detected in several solid tumors, including renal cell carcinoma (10) and gastric cancer (11). Moreover, Li et al. (12) reported that the positive expression rate of NLRC5 protein in patients with stage III non-small cell lung cancer (NSCLC) is close to 70%, and NSCLC patients with increased NLRC5 expression tend to present lower overall survival (OS) rate. It is worth mentioning that an increased level of NLRC5 protein has been found in HCC tissues by immunohistochemistry (IHC) in a study reported by He et al. (13). However, the expression of NLRC5 mRNA, the clinical significance of NLRC5, and the genes sets regulated by NLRC5 in HCC remain unclear.
In this study, we firstly obtained and analyzed information on NLRC5 mRNA expression in HCC from The Cancer Genome Atlas (TCGA) database and verified the results by performing quantitative real-time polymerase chain reaction (qRT-PCR) on the HCC tissues. Then, we measured the NLRC5 protein expression in HCC samples by conducting IHC and western blot analysis. Furthermore, we analyzed the clinical significance of NLRC5 in HCC after dividing the enrolled patients into NLRC5-positive and NLRC5-negative groups according to the IHC results. Finally, we conducted a gene set enrichment analysis (GSEA) to explore the gene sets regulated by NLRC5 in HCC.
Methods
Obtaining NLRC5 mRNA expression data from TCGA database
Published mRNA expression data related to HCC were downloaded from TCGA database (https://tcga-data.nci.nih.gov/tcga/). A total of 374 HCC samples and 50 non-cancer tissues were enrolled in TCGA. Limma package (version 3.8, http://www.bioconductor.org/packages/release/bioc/html/limma.html) (14) was used to extract NLRC5 expression data.
Clinical samples and animal resources
Samples used in this research were obtained from the Department of General Surgery, The East District of The First Affiliated Hospital of Anhui Medical University (also known as Feidong County People’s Hospital). The number of cases for qRT-PCR, western blot, and IHC, were 41, 32, and 60, respectively. All included volunteers were diagnosed as HCC by pathology. No patients had experienced chemotherapy or radiotherapy before surgery. OS was recorded from the date of operation to either the time of death or the date of the last follow-up. The clinical pathological characteristics of participants enrolled for IHC are listed in Table 1.
Table 1
| Clinical characteristic | Number | NLRC5 | χ2 | P | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Age (year) | 0.003 | 0.584 | |||
| ≤60 | 27 | 17 | 10 | ||
| >60 | 33 | 21 | 12 | ||
| Gender | 0.028 | 0.542 | |||
| Male | 39 | 25 | 14 | ||
| Female | 21 | 13 | 8 | ||
| Cirrhosis | 3.603 | 0.015 | |||
| With | 44 | 32 | 12 | ||
| Without | 16 | 6 | 10 | ||
| Tumor size (cm) | 1.279 | 0.194 | |||
| ≤5 | 27 | 15 | 12 | ||
| >5 | 33 | 23 | 10 | ||
| Number of tumors | 0.096 | 0.506 | |||
| Single | 45 | 28 | 17 | ||
| Multiple | 15 | 10 | 5 | ||
| Differentiation degree | 2.453 | 0.293 | |||
| High | 11 | 5 | 6 | ||
| Medium | 33 | 21 | 12 | ||
| Low | 16 | 12 | 4 | ||
| TNM stage | 6.969 | 0.011 | |||
| I+II | 39 | 20 | 19 | ||
| III+IV | 21 | 18 | 3 | ||
NLRC5, NLR family CARD domain containing 5; HCC, hepatocellular carcinoma.
This study was approved by the ethics committee of The East District of The First Affiliated Hospital of Anhui Medical University and conducted following the Declaration of Helsinki of 1975 (revised in 2008). All tissues were obtained with the consent of the patients and their families.
RNA extraction and qRT-PCR
RNA was extracted from 41 pairs of HCC and adjacent samples with TRIzol® reagent (Thermofisher, MA, USA; Cat. 15596018), which was then immediately reversed to cDNA. NLRC5 mRNA expression was measured with SensiFASTM SYBR No-ROX Kit (Bioline, London, UK, Cat. 98005). The qRT-PCR conditions were as follows: 95 °C for 2 minutes (1 cycle), 95 °C for 5 seconds (40 cycles), and 65 °C for 20 seconds (40 cycles). The value was normalized to GAPDH and calculated using the 2-ΔΔCq method. The forward and reverse primers for NLRC5 were ACAGCATCCTTAGACACTCCG and CCTTCCCCAAAAGCACGGT, respectively; for GAPDH, they were TGTGGGCATCAATGGATTTGG and ACACCATGTATTCCGGGTCAAT, respectively.
Western blotting assay
Total protein was extracted from 32 pairs of HCC and adjacent tissues using RIPA total protein lysate (Servicebio, Wuhan, China; Cat. G2002). The concentration of the obtained protein was then determined with a BCA protein assay kit (Servicebio, Wuhan, China, Cat. G2026). Protein was loaded into the SDS-PAGE gel and transferred to the membrane. The antibodies were added at a proper concentration after blocking. The primary antibodies for NLRC5 and GAPDH were anti-NLRC5 (Abcam, Cambridge, UK, Cat. Ab105411) and anti-GAPDH (Servicebio, Wuhan, China; Cat. GB12002), respectively.
IHC
The NLRC5 protein in 60 HCC and matched paracancerous samples were measured using IHC, as described previously by Cao et al. (15). The primary antibody for NLRC5 was anti-NLRC5 (Abcam, Cambridge, UK, Cat. Ab105411).
The total score of NLRC5 protein level was determined based on the product of the percentage of positive cells by staining intensity. More specifically, the percentage of positive cells was recorded as 1 point (0–25% staining), 2 points (26–50% staining), 3 points (51–75% staining), or 4 points (76–100% staining); staining intensity was recorded as 0 points (negative), 1 point (weak), 2 points (moderate), or 3 points (strong). The patients were then classified into an NLRC5-positive group (score >2, positive expression) or an NLRC5-negative group (score of 0–2).
GSEA enrichment analysis
GSEA software version 3.0 and high vs. low phenotype were employed for analysis (16). The nominal P value (Nom P value) <0.05 and the false discovery rate (FDR q-value) ≤0.25 were used as standards to select significantly changed gene sets. The data of the top 50 changed gene sets were exported. The ggplot2 packages (https://cran.r-project.org/web/packages/ggplot2/index.html) were used to merge graphs.
Statistical analysis
Beeswarm package (https://cran.r-project.org/web/packages/beeswarm/index.html) in R (version 3.5.1) and Mann-Whitney U test were used for analysis of data obtained from TCGA database. Furthermore, SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5 were employed for analysis of the experimental data. The Student’s t-test was used for the comparison of data in different groups. A P value <0.05 was considered statistically significant. Also, if the P value obtained by R and GSEA software was less than 0.0001, it was recorded in a scientific format.
Results
NLRC5 mRNA expression in HCC
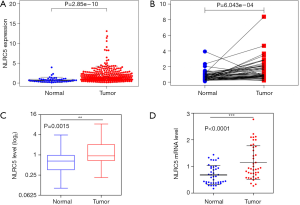
NLRC5 expression data obtained from TCGA database were analyzed, and the NLRC5 level was measured with qRT-PCR. Firstly, NLRC5 mRNA expression data in 374 HCC and 50 normal samples were analyzed, and a higher NLRC5 mRNA expression was found in tumor samples compared to normal ones (Figure 1A) (P=2.85e–10). Further, we matched the above unpaired samples, and 50 pairs of matched samples were obtained. By analyzing data of these paired cases using the U test and t-test, we found an enhanced NLRC5 mRNA expression in HCC samples (Figure 1B,C) (both P<0.05). Finally, qRT-PCR on 41 pairs of HCC and matched paracancerous tissues was conducted to verify the results obtained from TCGA database. We found that the qRT-PCR results were the same as the results obtained from the TCGA database (Figure 1D) (P<0.0001). In summary, data from TCGA database and qRT-PCR indicated that NLRC5 mRNA expression is higher in HCC tissues compared with that in paracancerous ones.
NLRC5 protein expression in HCC
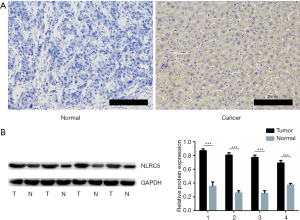
To measure the level of NLRC5 protein in HCC, IHC and western blot were performed on 60 and 32 paired HCC samples, respectively. IHC results revealed that the positive rates of NLRC5-positive cells in HCC and adjacent tissues were 63.3% (38/60) and 36.7% (22/60), respectively. As shown in Figure 2A, HCC tissues showed positive NLRC5 staining, while paracancerous tissues showed negative staining. The results of western blot were similar to those of IHC, with many more HCC tissues presenting higher NLRC5 protein levels (Figure 2B) (P<0.001). Overall, the NLRC5 protein level in HCC tissues exceeded that in paracancerous tissues.
The clinical significance of NLRC5 in HCC
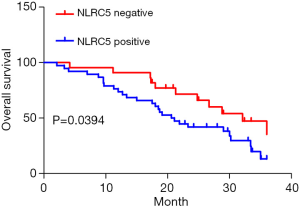
To investigate the clinical significance of NLRC5 in HCC, we classified the enrolled patients into 2 groups based on IHC results: the NLRC5-positive group and the NLRC5-negative group. A total of 38 patients with overall IHC score >2 were placed into the NLRC5-positive group, while the remaining 22 participants were placed into the NLRC5-negative group. By comparing the clinical pathological characteristics and 3-year OS rate of the 2 groups, we could determine the clinical significance of NLRC5 in HCC. As listed in Table 1, no significant difference could be found in age, gender, tumor size, and number of tumors between the 2 groups (all P>0.05). However, patients with a higher level of NLRC5 protein showed a higher rate of cirrhosis (P=0.015) and higher TNM stage (P=0.011). Moreover, the 3-year OS rates in the 2 groups varied: patients with a higher NLRC5 protein expression presented lower 3-year OS rate (Figure 3) (P=0.039). Therefore, the higher expression of NLRC5 was related to a higher rate of cirrhosis, higher TNM stage, and lower 3-year OS rate.
Gene sets regulated by NLRC5 in HCC
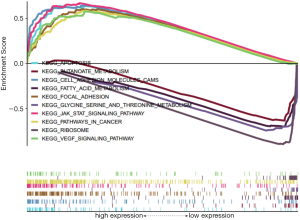
To explore gene sets regulated by NLRC5 in HCC, GSEA was performed based on the expression data obtained from TCGA database. A total of 177 gene sets were enrolled in the analysis. In the NLRC5-high phenotype, 131 gene sets were up-regulated, 98 of which were dramatically enriched at P<0.05, while in the NLRC5-low phenotype, 46 gene sets were up-regulated, 11 of which were markedly enriched at P<0.05. Of these changed gene sets, we selected a few typical sets to analyze with details of this analysis presented in Table 2 and Figure 4. Gene sets involved in the Janus kinases (JAK)/signal transducers and activators of transcription (STAT) signaling pathway, apoptosis, pathways in cancer, cell adhesion molecules, focal adhesion, and VEGF signaling pathway were up-regulated in the NLRC5-high phenotype; meanwhile, sets related to glycine serine and threonine metabolism, ribosome, butanoate metabolism, and fatty acid metabolism were up-regulated in the NLRC5-low phenotype.
Table 2
| Gene sets | NES | NOM p-val | FDR q-val |
|---|---|---|---|
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 2.23 | 0.00 | 0.00 |
| KEGG_APOPTOSIS | 2.06 | 0.00 | 3.21E−04 |
| KEGG_PATHWAYS_IN_CANCER | 2.06 | 0.00 | 3.02E−04 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 2.02 | 0.00 | 4.43E−04 |
| KEGG_FOCAL_ADHESION | 1.93 | 0.00 | 0.00 |
| KEGG_VEGF_SIGNALING_PATHWAY | 1.93 | 0.00 | 0.00 |
| KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM | −1.98 | 0.00 | 0.02 |
| KEGG_RIBOSOME | −1.84 | 0.00 | 0.04 |
| KEGG_BUTANOATE_METABOLISM | −1.78 | 0.02 | 0.07 |
| KEGG_FATTY_ACID_METABOLISM | −1.77 | 0.01 | 0.06 |
NLRC5, NLR family CARD domain containing 5; HCC, hepatocellular carcinoma.
Discussion
As a member of the NLR family, NLRC5 is involved in inflammasome activation and eventually causes the cleavage and activation of interleukin-18 (IL-18) and IL-1 (17). Another more commonly observed function of NLRC5 is as a transcriptional factor, and, in this role, NLRC5 serves as a transactivator of class I major histocompatibility complex (MHC I), which enables CD8+ T lymphocytes to identify and kill transformed cells (18-21). For instance, Li et al. (12) demonstrated that NLRC5 and MHC I expressions are positively correlated in 62 cases of NSCLC and 323 cases of 7 other different types of common human solid tumors. Also, recent studies have indicated that there is an abnormal expression of NLRC5 in several cancers, including renal cell carcinoma (10), gastric cancer (11), stage III NSCLC (12), and HCC (13). However, the expression of NLRC5 mRNA, the clinical significance of NLRC5, and the gene sets regulated by NLRC5 in HCC have remained relatively unexamined.
To explore this issue, we firstly compared NLRC5 expression in HCC and normal tissues using the data gathered from TCGA database and then verified the results obtained from TCGA database using qRT-PCR. Also, we measured the levels of the NLRC5 protein in the HCC samples using western blot and IHC. Moreover, the clinical significance of NLRC5 in HCC was analyzed. Finally, a GSEA was performed to investigate those gene sets regulated by NLRC5 in HCC.
Several studies have focused on NLRC5 expression and its relation to various cancers. Li et al. (12) reported a high NLRC5 expression in several solid cancers, including gastric and kidney cancer, cervical squamous cell carcinoma, gastric adenocarcinoma, malignant melanoma, and prostate and rectal cancer. Also, a higher expression of NLRC5 compared to paracancerous samples has been demonstrated in renal cell carcinoma, gastric cancer, stage III NSCLC, and HCC tissues (18-21). Moreover, the levels of NLRC5 were also found to be far higher in HCC and renal cell carcinoma cell lines than those in normal cell lines (18-21). The above results are consistent with the results of the present study. Briefly, we found that HCC tissues exhibited a higher NLRC5 mRNA and protein level compared to normal cases. However, a study on colorectal cancer conducted by Liu et al. indicated no marked difference in the NLRC5 expression between cancerous and normal tissues (22).
The clinical significance of NLRC5 in renal cell carcinoma, gastric cancer, stage III NSCLC, and colorectal cancer has been well-documented. Li et al. (11) showed that NLRC5 protein level is related to the site of the tumor, the number of lymph nodes, and the TNM stage in gastric cancer and to the OS rate in gastric cancer patients. Similarly, Wang et al. (10) reported that patients with higher NLRC5 expression are more likely to present a lower OS and higher TNM stage. Also, Li et al. (12) concluded that higher nuclear NLRC5 is associated with lower OS. In addition, a study on NLRC5 polymorphism in colorectal cancer showed that patients with the minor alleles of rs27194 and rs289747, which are two variants of NLRC5, showed lower OS rate (23). However, the clinical significance of NLRC5 in HCC is still unknown. In this study, we classified the recruited HCC patients into NLRC5-positive and NLRC5-negative groups based on the IHC result. Through comparing the clinicopathological characteristics and OS rate in the 2 groups, we concluded that patients with higher NLRC5 levels are related to a higher rate of cirrhosis, higher TNM stage, and lower 3-year OS rate.
Few studies on the function and mechanism of NLRC5 in controlling cancer progression have been performed. He et al. (13) found that silencing NLRC5 inhibits the proliferation of HCC cell lines (HepG2) by causing a significant drop in the level of protein kinase B (AKT) and a marked increase of VEGF-A. Also, knocking down NLRC5 in renal cell carcinoma cell lines (786-O and 769-P) weakened the proliferation and metastasis of these cells via down-regulating the expression of β-catenin, c-Myc, and cyclin D1 (10). We found that NLRC5 up-regulated the JAK/STAT pathway and gene sets associated with apoptosis. STAT has also been reported to regulate the level of various proteins related to the induction or prevention of apoptosis (24). Hence, NLRC5 may be involved in the regulation of apoptosis of HCC through the JAK/STAT pathway. Also, we found that NLRC5 up-regulated gene sets involved in cell adhesion molecules, focal adhesion, and VEGF signaling pathway. The VEGF family can promote tumor metastasis by affecting the neovascularization of blood vessels and lymphatics (25,26). Furthermore, cell adhesion and focal adhesion are 2 vital components in tumor metastasis. Therefore, NLRC5 can promote HCC metastasis.
Some limitations of this study should be addressed. First, the number of samples recruited for the experiment were limited. Second, the clinical roles of NLRC5 in HCC in TCGA database have not been analyzed. Third, experiments studying the roles and mechanisms of NLRC5 in HCC progression have yet to be conducted, and studies on mice that could further verify our results are lacking. Therefore, further related experiments should be performed to address the above limitations.
In summary, our study found that NLRC5 levels were increased in HCC tissues, and an enhanced NLRC5 expression was related to a higher rate of cirrhosis, a higher TNM stage, and lower 3-year OS rate. Moreover, gene sets related to the metastasis of cancer were up-regulated by NLRC5 in HCC. Therefore, NLRC5 may be a valuable marker for the diagnosis and prognostic assessment of HCC.
Acknowledgments
We would like to thank our colleagues in The First Affiliated Hospital of Soochow University and The East District of First Affiliated Hospital of Anhui Medical University who gave me advisable suggestion on this project.
Funding: None.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure from (available at http://dx.doi.org/10.21037/tcr.2020.02.81). The authors have no conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the ethics committee of The East District of The First Affiliated Hospital of Anhui Medical University and conducted following the Declaration of Helsinki of 1975 (revised in 2008). All tissues were obtained with the consent of the patients and their families.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vogel A, Saborowski A. Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat Rev 2020;82:101946. [Crossref] [PubMed]
- Lim LJ, Wong SYS, Huang F, et al. Roles and Regulation of Long non-coding RNAs in Hepatocellular Carcinoma. Cancer Res 2019;79:5131-9. [Crossref] [PubMed]
- Zhou F, Shang W, Yu X. Med Res Rev 2018;38:741-67. [Crossref] [PubMed]
- Guan L, Wu W, Pang H, et al. Anti-GPC3 single-chain scFv antibody acts as an agent for radio-immunoimaging in diagnosing hepatocellular carcinoma. Am J Transl Res 2019;11:7422-31. [PubMed]
- Adachi H, Derevnina L, Kamoun S. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr Opin Plant Biol 2019;50:121-31. [Crossref] [PubMed]
- Wu X, Dong L, Lin X, et al. Relevance of the NLRP3 Inflammasome in the Pathogenesis of Chronic Liver Disease. Front Immunol 2017;8:1728. [Crossref] [PubMed]
- Tartey S, Kanneganti TD. Differential role of the NLRP3 inflammasome in infection and tumorigenesis. Immunology 2019;156:329-38. [Crossref] [PubMed]
- Velloso FJ, Trombetta-Lima M, Anschau V, et al. NOD-like receptors: major players (and targets) in the interface between innate immunity and cancer. Biosci Rep 2019; [Crossref] [PubMed]
- Tong Y, Cui J, Li Q, et al. Enhanced TLR-induced NF-kappaB signaling and type I interferon responses in NLRC5 deficient mice. Cell Res 2012;22:822-35. [Crossref] [PubMed]
- Wang Q, Ding H, He Y, et al. NLRC5 mediates cell proliferation, migration, and invasion by regulating the Wnt/beta-catenin signalling pathway in clear cell renal cell carcinoma. Cancer Lett 2019;444:9-19. [Crossref] [PubMed]
- Li Y, Zhang M, Zheng X. High Expression of NLRC5 Is Associated with Prognosis of Gastric Cancer. Open Med (Wars) 2018;13:443-9. [Crossref] [PubMed]
- Li X, Guo F, Liu Y, et al. NLRC5 expression in tumors and its role as a negative prognostic indicator in stage III non-small-cell lung cancer patients. Oncol Lett 2015;10:1533-40. [Crossref] [PubMed]
- He YH, Li MF, Zhang XY, et al. NLRC5 promotes cell proliferation via regulating the AKT/VEGF-A signaling pathway in hepatocellular carcinoma. Toxicology 2016;359-360:47-57. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Cao GD, Chen K, Chen B, et al. Positive prognostic value of HER2-HER3 co-expression and p-mTOR in gastric cancer patients. BMC Cancer 2017;17:841. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 2015;21:677-87. [Crossref] [PubMed]
- Davis BK, Roberts RA, Huang MT, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol 2011;186:1333-7. [Crossref] [PubMed]
- Meissner TB, Li A, Biswas A, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A 2010;107:13794-9. [Crossref] [PubMed]
- Chelbi ST, Guarda G. NLRC5, a promising new entry in tumor immunology. J Immunother Cancer 2016;4:39. [Crossref] [PubMed]
- Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol 2012;12:813-20. [Crossref] [PubMed]
- Liu R, Truax AD, Chen L, et al. Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components. Oncotarget 2015;6:33456-69. [PubMed]
- Catalano C, Da SFM, Jiraskova K, et al. Short article: Influence of regulatory NLRC5 variants on colorectal cancer survival and 5-fluorouracil-based chemotherapy. Eur J Gastroenterol Hepatol 2018;30:838-42. [Crossref] [PubMed]
- Bousoik E, Montazeri AH. "Do We Know Jack" About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front Oncol 2018;8:287. [Crossref] [PubMed]
- Eisermann K, Fraizer G. The Androgen Receptor and VEGF: Mechanisms of Androgen-Regulated Angiogenesis in Prostate Cancer. Cancers (Basel) 2017; [Crossref] [PubMed]
- Simon T, Gagliano T, Giamas G. Direct Effects of Anti-Angiogenic Therapies on Tumor Cells: VEGF Signaling. Trends Mol Med 2017;23:282-92. [Crossref] [PubMed]


