
Screening key lncRNAs and mRNAs for left-sided and right-sided colon adenocarcinoma based on lncRNA-mRNA functional synergistic network
Introduction
Colon cancer is the fourth most malignant tumor worldwide and the fifth leading cause of cancer-related death (1). A growing body of studies suggests that right-sided colon adenocarcinoma (RSCOAD) and left-sided colon adenocarcinoma (LSCOAD) should be recognized as two distinct categories of cancer (2,3). Recently, the differences between RSCOAD and LSCOAD have attracted people’s attention due to their different outcomes, prognoses, and clinical responses to chemotherapy (2). Primary tumor location associates with survival in patients with metastatic COAD (4). Therefore, it has become an urgent tissue to find the key molecular markers of RSCOAD and LSCOAD.
With the development of gene expression profiles, bioinformatics have become most common used strategies to screen key biomarkers in a variety of diseases (5-7). The miRNAs associated with RSCOAD and LSCOAD has been reported in our previous study (8). In recent years, a large number of evidence suggests that abnormal expression of lncRNA contributes to the development of human cancer (9-11). LncRNA plays a vital role in biological processes including cancer cells proliferation, metastasis and apoptosis (12,13). However, research for lncRNA underlying biomarkers in RSCOAD and LSCOAD is rarely. Thence, screening for pivotal lncRNAs is essential for understanding pathological mechanism of RSCOAD and LSCOAD. In this study, we obtained the lncRNA and mRNA expression data of RSCOAD and LSCOAD patients from TCGA, and tried to identify the optimal diagnostic lncRNA biomarkers using Boruta algorithm. The differentially expressed mRNA (DEmRNA)-differentially expressed lncRNA (DElncRNA) interaction analysis was performed to uncover the key DElncRNA in RSCOAD and LSCOAD. The qRT-PCR was applied to validate the candidate DEmRNAs and DElncRNAs. To our knowledge, this is the first time to find key lncRNAs in RSCOAD and LSCOAD using random forest model.
Methods
Integrated profiles in The Cancer Genome Atlas (TCGA)
In this study, the lncRNA and mRNA gene expression profiles and clinical data of RSCOAD and LSCOAD was download from TCGA (http://tcga-data.nci.nih.gov/). The inclusion criteria for the present study were: (I) histological type is COAD. (II) Anatomic neoplasm subdivision type includes ascending colon, sigmoid colon, cecum and descending colon.
Identification DElncRNAs and DEmRNAs between RSCOAD and LSCOAD
We filtered and deleted the undetectable lncRNAs and mRNAs (with read count value =0 in more than 20% RSCOAD case or in more than 20% LSCOAD). The DElncRNAs and DEmRNAs between RSCOAD and LSCOAD was calculate using the R-Bioconductor package DESeq2, and the P value was then calculated. Multiple comparisons were performed by the Benjamini and Hochberg approach, and the false discovery rate (FDR) was then obtain the. DElncRNAs and DEmRNAs were defined with the thresholds of FDR <0.01. The R package was used to perform the hierarchical clustering analysis of DElncRNAs and DEmRNAs.
Features selection
Feature selection can readily remove redundant and irrelevant features, thereby further improving the performance of a classifier. Boruta algorithm (https://cran.r-project.org/web/packages/Boruta/) was used to minimize errors of random forest model and further obtain an optimal feature subset. In the algorithm of Boruta, we used the Z-score as measurement criteria.
DEmRNA-DElncRNA interaction analysis
To identify the DEmRNAs near DElncRNAs with cis-regulatory effects, DEmRNAs transcribed within a 100 kb window up- or down-stream of DElncRNAs in RSCOAD and LSCOAD were identified. The DEmRNAs co-expressed with DElncRNAs were identified as well. Pairwise Pearson correlation coefficient between DEmRNAs and DElncRNAs were calculated. DElncRNA-DEmRNA pairs with |r|>0.7 and P<0.05 were defined as significant mRNA-lncRNA co-expression pairs. The DEmRNA-DElncRNA interaction network were construct by the Cytoscape software (http://www.cytoscape.org/).
Functional annotation
To uncover the biological functions of the DEmRNA co-expressed with DElncRNA, GO classification and KEGG pathway enrichment analysis were executed using Metascape (http://metascape.org/gp/index.html). A FDR <0.05 was defined as the criteria of statistical significance.
Survival analysis
By using the survival analysis (https://cran.r-project.org/web/packages/survival/index.html) in R, we analyzed the association between clinical factors and the survival rate of RSCOAD and LSCOAD patients. Univariate Cox regression analysis was performed for each clinical factor. Multivariate Cox regression analysis was conducted for survival factors obtained by univariate Cox regression. P<0.05 was considered statistically significant.
Confirmation by qRT-PCR
Fourteen tissues samples of RSCOAD patients (n=7) and LSCOAD patients (n=7) were obtained. Informed written consent was obtained from all participants. The study was approved by institutional ethics committee of Nankai Hospital of Tianjin [No. NKYY_YXKT(B)_IRB_2019_101_01].
The qRT-PCR was performed as previously reported (8). The PCR primers used are listed in Table 1.
Table 1
| Name | Sequence (5' to 3') |
|---|---|
| GAPDH-F | GGAGCGAGATCCCTCCAAAAT |
| GAPDH-R | GGCTGTTGTCATACTTCTCATGG |
| CYP4F2-F | GAGGGTAGTGCCTGTTTGGAT |
| CYP4F2-R | CAGGAGGATCTCATGGTGTCTT |
| HOXB3-F | CCAGTGCCACTAGCAACAG |
| HOXB3-R | CGTTTGCCTCGACTCTTTCATC |
| HOXD1-F | CGGGTCTCACGTCCACTAC |
| HOXD1-R | GATGCGGTCTGGAAAGCAC |
| UCA1-F | CGGACATGCTTGACACTTGGT |
| UCA1-R | CAGTCTTCAGCCACTAAGCCG |
| HOXB-AS3-F | AAGTAGAGCCTCCACGACCC |
| HOXB-AS3-R | GAGGAAACGGCCGGTCAATC |
| HAGLR-F | GATTTGGTCCAAGCCCTCACC |
| HAGLR-R | AGTGTCATTTGCGGCTTAGGG |
Results
DEmRNAs and DElncRNAs between RSCOAD and LSCOAD
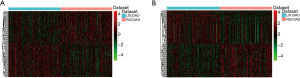
The clinical data of 156 RSCOAD and 158 LSCOAD patients were shown in Table 2. We obtained the mRNA and lncRNA expression profiles of 156 RSCOAD and 158 LSCOAD patients from TCGA. A total of 2,672 DEmRNAs (1,050 down-regulated and 1,622 up-regulated mRNAs) and 453 DElncRNAs (139 down-regulated and 314 up-regulated lncRNAs) between RSCOAD and LSCOAD were identified with an FDR <0.01. Hierarchical clustering analysis of the top 100 DElncRNAs and DEmRNAs are displayed in Figure 1, respectively.
Table 2
| Clinicopathological parameters | RSCOAD [156] | LSCOAD [158] | Total [314] | P |
|---|---|---|---|---|
| Age | 0.001 | |||
| Mean ± SD | 69.391±12.473 | 64.899±12.095 | 67.131±12.469 | |
| Median | 71 | 66 | 68 | |
| Gender | 0.644 | |||
| Female | 69 | 75 | 144 | |
| Male | 87 | 83 | 170 | |
| Weight | 0.869 | |||
| Mean ± SD | 81.365±21.508 | 81.907±19.990 | 81.608±20.780 | |
| Median | 79.6 | 78.25 | 78.9 | |
| NA | 67 | 86 | 153 | |
| Race | 0.135 | |||
| White | 72 | 66 | 138 | |
| Black or African American | 28 | 12 | 40 | |
| ASIAN | 4 | 3 | 7 | |
| NA | 52 | 77 | 129 | |
| Lymph node count | 0.013 | |||
| Mean ± SD | 23.660±11.017 | 22.164±15.642 | 22.900±13.565 | |
| Median | 22 | 18 | 20 | |
| NA | 9 | 6 | 15 | |
| Lymphatic invasion | 0.113 | |||
| Yes | 52 | 67 | 119 | |
| No | 88 | 75 | 163 | |
| NA | 16 | 16 | 32 | |
| Number of positive lymph node | 0.196 | |||
| Mean ± SD | 2.243±5.531 | 1.914±3.912 | 2.075±4.766 | |
| Median | 0 | 0 | 0 | |
| NA | 12 | 7 | 19 | |
| Neoplasm cancer status | 0.114 | |||
| With tumor | 68 | 86 | 154 | |
| Tumor free | 69 | 58 | 127 | |
| NA | 19 | 14 | 33 | |
| Preoperative pretreatment CEA level | 0.290 | |||
| Mean ± SD | 43.619±267.729 | 52.469±197.436 | 47.977±235.197 | |
| Median | 2.8 | 3.175 | 2.93 | |
| NA | 55 | 60 | 115 | |
| Residual tumor | 0.105 | |||
| R0 | 112 | 116 | 228 | |
| R1 | 2 | 0 | 2 | |
| R2 | 6 | 14 | 20 | |
| RX | 10 | 6 | 16 | |
| NA | 26 | 22 | 48 | |
| Stage | 0.176 | |||
| Stage I | 29 | 26 | 55 | |
| Stage II | 65 | 53 | 118 | |
| Stage III | 34 | 49 | 83 | |
| Stage IV | 21 | 28 | 49 | |
| NA | 7 | 2 | 9 | |
| M | 0.154 | |||
| M0 | 111 | 118 | 229 | |
| M1 | 21 | 28 | 49 | |
| MX | 20 | 11 | 31 | |
| NA | 4 | 1 | 5 | |
| N | 0.011 | |||
| N0 | 101 | 84 | 185 | |
| N1 | 25 | 48 | 73 | |
| N2 | 30 | 26 | 56 | |
| T | 0.687 | |||
| T1 | 5 | 4 | 9 | |
| T2 | 28 | 31 | 59 | |
| T3 | 103 | 109 | 212 | |
| T4 | 20 | 14 | 34 |
RSCOAD, right-sided colon adenocarcinoma; LSCOAD, left-sided colon adenocarcinoma; CEA, carcinoembryonic antigen.
Features selection
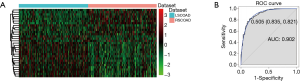
We obtained 31 DElncRNAs using algorithms of Boruta (Table 3). Hierarchical clustering analysis of these 31 DElncRNAs between RSCOAD and LSCOAD are shown in Figure 2A. The 10-fold cross-validation result demonstrated that the AUC of the random forests model was 0.902, and the specificity and sensitivity of this model were 83.5% and 82.1%, respectively (Figure 2B).
Table 3
| Symbol | baseMean | log2 (fold change) | P value | FDR | Up/down |
|---|---|---|---|---|---|
| UNC5B-AS1 | 1.470E+01 | 1.467E+00 | 3.186E–17 | 1.151E–13 | Up |
| SNHG11 | 4.832E+02 | –5.342E–01 | 2.056E–14 | 1.857E–11 | Down |
| HAGLR | 3.513E+02 | 1.078E+00 | 1.830E–14 | 1.857E–11 | Up |
| AC005256.1 | 1.320E+01 | 1.400E+00 | 1.890E–14 | 1.857E–11 | Up |
| RP11-9E17.1 | 5.870E+01 | 8.566E–01 | 1.165E–13 | 8.417E–11 | Up |
| RP5-881L22.5 | 3.003E+02 | –8.667E–01 | 7.642E–13 | 3.945E–10 | Down |
| RP1-101A2.1 | 3.533E+01 | –6.150E–01 | 2.196E–12 | 8.818E–10 | Down |
| TFAP2A-AS1 | 2.585E+01 | 1.104E+00 | 6.223E–12 | 2.045E–09 | Up |
| LINC01315 | 1.737E+02 | –6.322E–01 | 2.334E–11 | 5.658E–09 | Down |
| FEZF1-AS1 | 1.406E+02 | 1.228E+00 | 1.215E–10 | 1.996E–08 | Up |
| ZNF528-AS1 | 3.894E+01 | –8.155E–01 | 2.007E–10 | 2.790E–08 | Down |
| RP4-647C14.2 | 1.232E+01 | 7.463E–01 | 2.709E–09 | 2.577E–07 | Up |
| RP11-431J24.2 | 1.508E+02 | –1.141E+00 | 1.663E–08 | 1.279E–06 | Down |
| RP11-473M20.9 | 1.007E+02 | –8.270E–01 | 2.060E–08 | 1.520E–06 | Down |
| B3GALT5-AS1 | 2.111E+01 | –1.098E+00 | 2.158E–08 | 1.549E–06 | Down |
| LA16c-313D11.12 | 6.570E+01 | 5.271E–01 | 6.377E–08 | 3.906E–06 | Up |
| RP11-680F8.1 | 6.675E+01 | –9.165E–01 | 8.883E–08 | 4.864E–06 | Down |
| AC007277.3 | 4.094E+01 | 1.015E+00 | 1.637E–07 | 7.994E–06 | Up |
| RP11-395B7.2 | 3.198E+02 | –6.241E–01 | 1.891E–07 | 8.759E–06 | Down |
| AC106876.2 | 1.899E+02 | –5.087E–01 | 3.051E–07 | 1.253E–05 | Down |
| SATB2-AS1 | 4.174E+02 | –6.758E–01 | 2.507E–06 | 7.025E–05 | Down |
| HOXB-AS3 | 2.991E+02 | 6.179E–01 | 2.759E–06 | 7.612E–05 | Up |
| RP11-834C11.4 | 3.131E+01 | 5.172E–01 | 1.428E–05 | 2.837E–04 | Up |
| AC022182.3 | 1.469E+01 | 7.504E–01 | 1.777E–05 | 3.399E–04 | Up |
| LINC01558 | 1.053E+02 | –5.200E–01 | 5.458E–05 | 8.496E–04 | Down |
| UCA1 | 9.408E+02 | –6.980E–01 | 6.137E–05 | 9.280E–04 | Down |
| AP003774.1 | 3.259E+02 | –6.402E–01 | 6.305E–05 | 9.456E–04 | Down |
| AC011298.2 | 4.616E+01 | –6.174E–01 | 1.141E–04 | 1.516E–03 | Down |
| DIO3OS | 5.232E+02 | –5.439E–01 | 1.341E–04 | 1.730E–03 | Down |
| CRAT8 | 5.560E+01 | –4.839E–01 | 5.627E–04 | 5.438E–03 | Down |
| LINC01485 | 1.901E+01 | –5.874E–01 | 1.014E–03 | 8.525E–03 | Down |
The bold mark is top 10 up/down. FDR, false discovery rate.
DElncRNA-DEmRNA co-expression analysis
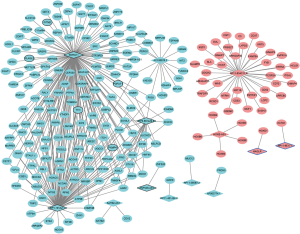
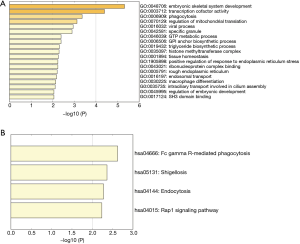
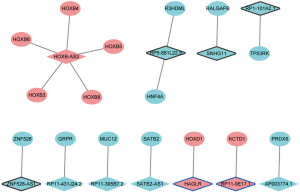
A total of 343 DElncRNA-DEmRNA co-expression pairs including 13 DElncRNAs and 230 DEmRNAs were identified with absolute value of the Pearson correlation coefficient |r|>0.7 and P<0.05. The co-expressed DElncRNA-DEmRNA network (Figure 3) was consisted of 243 nodes and 343 edges. Among which, SNHG11 (degree =161), RP1-101A2.1 (degree =95), RP5-881L22.5 (degree =20) and HAGLR (degree =1) were top 10 DElncRNA. We also performed the functional annotation of 230 DEmRNAs co-expressed with DElncRNAs. The GO enrichment and KEGG enrichment results are shown in Figure 4.
DElncRNAs-nearby DEmRNAs interaction network
Considering that the functions of most lncRNAs have been unknown. We speculated that the lncRNAs may play roles by regulating their nearby genes. A total of 39 DElncRNAs-nearby target DEmRNAs pairs were obtained which were consisted of 21 DElncRNAs and 40 DEmRNAs (Figure 5). After overlapped DElncRNAs-DEmRNAs co-expression network with DElncRNAs-nearby DEmRNAs interaction network, we obtained a total of 16 DElncRNA-DEmRNA pairs including 11 DElncRNAs and 16 DEmRNAs (Figure 6).
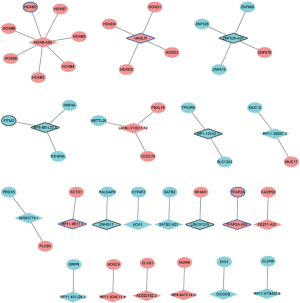
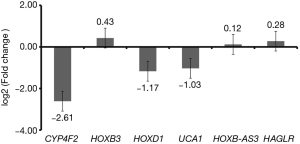
qRT-PCR confirmation
We performed the confirmation of candidate DEmRNAs (CYP4F2, HOXB3 and HOXD1) and DElncRNAs (UCA1, HOXB-AS3 and HAGLR) using qRT-PCR. Based on TCGA, CYP4F2 and UCA1 were down-regulated while the other four DEmRNAs or DElncRNAs (HOXB3, HOXD1, HOXB-AS3 and HAGLR) were up-regulated in RSCOAD compared to LSCOAD. According to the qRT-PCR results, except for HOXD1, CYP4F2 and UCA1 were down-regulated and HOXB3, HOXB-AS3 and HAGLR were up-regulated which were consistent with the results of TCGA, generally (Figure 7).
Regression Analysis of univariate Cox and multivariate Cox
A univariate Cox regression analysis showed that age, lymphatic invasion, neoplasm status, carcinoembryonic antigen (CEA) level, residual tumor, stage and M were associated with survival (Table 4). The survival curves results of age, neoplasm status, CEA level, residual tumor and M had a significant prognostic value (Figure 8A,B,C,D,E). As shown in Figure 8F, the overall prognosis of RSCOAD was worse than that of LSCOAD, but there is no significant difference. We also performed the multivariate Cox regression analysis, and results showed that age, residual tumor, stage, and M were independent predictors of survival (Table 5).
Table 4
| Parameters | coef | exp(coef) | Lower 0.95 | Upper 0.95 | se(coef) | z | Pr(>|z|) |
|---|---|---|---|---|---|---|---|
| Tumor position | 2.860E–01 | 1.331E+00 | 6.848E–01 | 2.587E+00 | 3.391E–01 | 8.433E–01 | 3.990E–01 |
| Age | 3.135E–02 | 1.032E+00 | 1.004E+00 | 1.061E+00 | 1.414E–02 | 2.217E+00 | 2.659E–02 |
| Male | –4.265E–01 | 6.528E–01 | 3.314E–01 | 1.286E+00 | 3.458E–01 | –1.233E+00 | 2.174E–01 |
| Weight | –1.731E–02 | 9.828E–01 | 9.506E–01 | 1.016E+00 | 1.702E–02 | –1.017E+00 | 3.089E–01 |
| Race | –1.681E–01 | 8.453E–01 | 2.869E–01 | 2.491E+00 | 5.513E–01 | –3.048E–01 | 7.605E–01 |
| Lymph node count | –2.751E–02 | 9.729E–01 | 9.407E–01 | 1.006E+00 | 1.717E–02 | –1.602E+00 | 1.091E–01 |
| Number of positive lymph node | 1.801E–02 | 1.018E+00 | 9.743E–01 | 1.064E+00 | 2.247E–02 | 8.017E–01 | 4.227E–01 |
| Lymphatic invasion | –8.312E–01 | 4.355E–01 | 2.114E–01 | 8.973E–01 | 3.688E–01 | –2.254E+00 | 2.420E–02 |
| Neoplasm cancer status | 2.682E+00 | 1.461E+01 | 4.313E+00 | 4.949E+01 | 6.225E–01 | 4.308E+00 | 1.648E–05 |
| CEA level | 8.261E–04 | 1.001E+00 | 1.000E+00 | 1.002E+00 | 4.174E–04 | 1.979E+00 | 4.778E–02 |
| Residual tumor | 1.703E+00 | 5.489E+00 | 2.004E+00 | 1.503E+01 | 5.140E–01 | 3.313E+00 | 9.242E–04 |
| Stage | 8.785E–01 | 2.407E+00 | 1.194E+00 | 4.853E+00 | 3.577E–01 | 2.456E+00 | 1.406E–02 |
| M | 9.741E–01 | 2.649E+00 | 1.329E+00 | 5.279E+00 | 3.518E–01 | 2.769E+00 | 5.626E–03 |
| N | 6.513E–01 | 1.918E+00 | 9.925E–01 | 3.706E+00 | 3.361E–01 | 1.938E+00 | 5.266E–02 |
| T | 3.300E–01 | 1.391E+00 | 4.851E–01 | 3.988E+00 | 5.374E–01 | 6.141E–01 | 5.392E–01 |
CEA, carcinoembryonic antigen.
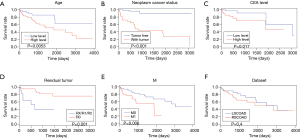
Table 5
| Parameters | coef | exp(coef) | Lower 0.95 | Upper 0.95 | se(coef) | z | Pr(>|z|) |
|---|---|---|---|---|---|---|---|
| Age | 1.780E–01 | 1.195E+00 | 1.017E+00 | 1.403E+00 | 8.206E–02 | 2.169E+00 | 3.006E–02 |
| Lymphatic invasion | 9.473E–01 | 2.579E+00 | 2.562E–01 | 2.595E+01 | 1.178E+00 | 8.041E–01 | 4.213E–01 |
| Neoplasm cancer status | 2.052E+01 | 8.172E+08 | 0.000E+00 | Inf | 3.828E+03 | 5.361E–03 | 9.957E–01 |
| CEA level | 1.280E–03 | 1.001E+00 | 9.992E–01 | 1.003E+00 | 1.071E–03 | 1.196E+00 | 2.319E–01 |
| Residual tumor | 2.684E+00 | 1.464E+01 | 1.242E+00 | 1.726E+02 | 1.259E+00 | 2.132E+00 | 3.301E–02 |
| Stage | 5.794E+00 | 3.284E+02 | 4.955E+00 | 2.177E+04 | 2.140E+00 | 2.708E+00 | 6.771E–03 |
| M | –4.528E+00 | 1.081E–02 | 1.546E–04 | 7.553E–01 | 2.167E+00 | –2.089E+00 | 3.667E–02 |
CEA, carcinoembryonic antigen.
Discussion
The distinction between RSCOAD and LSCOAD has been a serious dispute for a long time. Therefore, a comprehensive and detailed study is crucial to reveal the differences between RSCOAD and LSCOAD. In this study, the lncRNA and mRNA gene expression profiles were obtained in patients with RSCOAD and LSCOAD from TCGA. A total of 2,672 DEmRNAs (1,050 down-regulated and 1,622 up-regulated mRNAs) and 453 DElncRNAs (139 down-regulated and 314 up-regulated lncRNAs) between RSCOAD and LSCOAD were identified. Additionally, 31 DElncRNAs between RSCOAD and LSCOAD were identified by the algorithms of Boruta. Moreover, we build the DElncRNA-DEmRNA interaction network to find pivotal DEmRNAs and DElncRNAs, and also performed the survival analysis. Finally, the expression of selected DEmRNAs and DElncRNAs were verified using qRT-PCR. HAGLR acts as a microRNA-143-5p sponge to modulate epithelial-mesenchymal transition and metastatic potential in esophageal cancer through modulating LAMP3 (14). HAGLR inhibited cell proliferation of lung cancer by epigenetically silencing E2F1 (15). HAGLR is up-regulated in osteosarcoma and non-small cell lung cancer, and it is involved in the development and poor prognosis of these cancers (16,17). In the current study, HAGLR was down-regulated in both TCGA integration analysis and qRT-PCR validation, indicating the TCGA integration analysis data were reliable. Results of DElncRNA-DEmRNA co-expression analysis showed that HAGLR was co-expressed with HOXD1. Therefore, we speculated that HOXD1 might associated with the progression of RSCOAD and LSCOAD by regulating HAGLR.
Huang et al. reported that HOXB-AS3 (HOXB cluster antisense RNA 3) encodes a conserved 53-aa peptide, and the peptide plays crucial role in colon cancer growth (18). In this study, HOXB-AS3 was co-expressed with HOXB6, HOXB5, HOXB4, HOXB3 and HOXB8. HOXB6 might involve in the development of colorectal cancer (19). The risk score of HOXB3 gene expression may be helpful for stratification of prognostic risk in high-grade serous ovarian cancer patients, and provides a basis for prospective verification (20). MiR-375 suppresses cancer stem cell phenotype and tamoxifen resistance via degrading HOXB3 in human ER-positive breast cancer (21). HOXB3 is involved in regulating colon cancer cell proliferation and invasion, and HOXB3 is a potential target for colon cancer therapy (22). Herein, HOXB3 was up-regulated in our TCGA integration analysis and qRT-PCR validation, which was consistent with reports in other cancer suggesting the results were convincing. Therefore, we further hypothesized that HOXB-AS3 might play key roles in RSCOAD and LSCOAD by regulating HOXB3.
SATB2, a transcription factor involved in chromatin remodeling, has been identified as a novel immunohistochemistry marker with relatively high sensitivity for colorectal carcinoma (23). SATB2 is a specific immunohistochemistry marker that can be used to diagnose metastatic and primary signet ring cell carcinomas of lower gastrointestinal origin (24). LncRNA SATB2-AS1 suppresses tumor metastasis of colorectal cancer and regulates the microenvironment of tumor immune cells through regulating SATB2 (25). In this study, we found that SATB2 was down-regulated in both TCGA integration analysis. Results of DElncRNA-DEmRNA co-expression analysis showed that SATB2-AS1 was co-expressed with SATB2. Therefore, we speculated that SATB2-AS1 might involve in the progression of RSCOAD and LSCOAD by regulating SATB2.
RSCOAD patients have a poor survival than LSCOAD patients (26). In the study, cox regression analysis showed that the overall prognosis of RSCOAD was worse than that of LSCOAD, which was consistent with other researcher reports, suggesting our TCGA integration results were reliable.
In conclusion, we obtained 2,672 DEmRNAs and 45 DElncRNAs in RSCOAD compared to LSCOAD. The Boruta algorithm was to identify 31 optimal diagnostic lncRNAs biomarkers between RSCOAD and LSCOAD. Among them, three DElncRNAs (HAGLR, HOXB-AS3 and SATB2-AS1) and three DEmRNAs (HOXD1, HOXB3 and SATB2) were identified pivotal DElncRNAs and DEmRNAs between RSCOAD and LSCOAD. Some limitations of our study should be mentioned. More samples are needed to validate expression of pivotal DElncRNAs and DEmRNAs. In addition, in vivo and in vitro experiments are necessary to uncover the biological functions of key DElncRNAs and DEmRNAs in RSCOAD and LSCOAD in future study.
Acknowledgments
We thank Beijing Medintell Bioinformatic Technology Co., LTD for assistance in high-throughput sequencing and data analysis.
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by institutional ethics committee of Nankai Hospital of Tianjin [No. NKYY_YXKT(B)_IRB_2019_101_01]. Informed written consent was obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Kim K, Kim YW, Shim H, et al. Differences in clinical features and oncologic outcomes between metastatic right and left colon cancer. J BUON 2018;23:11-8. [PubMed]
- Jensen CE, Villanueva JY, Loaiza-Bonilla A. Differences in overall survival and mutation prevalence between right- and left-sided colorectal adenocarcinoma. J Gastrointest Oncol 2018;9:778-84. [Crossref] [PubMed]
- Ahmed S, Pahwa P, Le D, et al. Primary tumor location and survival in the general population with metastatic colorectal cancer. Clin Colorectal Cancer 2018;17:e201-6. [Crossref] [PubMed]
- Tao L, Yang L, Huang X, et al. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in dilated cardiomyopathy. Front Genet 2019;10:1149. [Crossref] [PubMed]
- Zhou D, Gao B, Yang Q, et al. Integrative analysis of ceRNA network reveals functional lncRNAs in intrahepatic cholangiocarcinoma. Biomed Res Int 2019;2019:2601271.
- Wang Y, Wu N, Liu J, et al. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol 2015;10:131. [Crossref] [PubMed]
- Yang L, Li L, Ma J, et al. miRNA and mRNA integration network construction reveals novel key regulators in left-sided and right-sided colon adenocarcinoma. Biomed Res Int. 2019;2019:7149296.
- Wang Y, Chen S, Chen L, et al. Associating lncRNAs with small molecules via bilevel optimization reveals cancer-related lncRNAs. PLoS Comput Biol 2019;15:e1007540. [Crossref] [PubMed]
- Li H, Gao C, Liu L, et al. 7-lncRNA assessment model for monitoring and prognosis of breast cancer patients: based on cox regression and co-expression analysis. Front Oncol 2019;9:1348. [Crossref] [PubMed]
- Wang J, Zhang C, Wu Y, et al. Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in bladder urothelial carcinoma. Cancer Cell Int 2019;19:327. [Crossref] [PubMed]
- Liu S, Li J, Kang L, et al. Degradation of long non-coding RNA-CIR decelerates proliferation, invasion and migration, but promotes apoptosis of osteosarcoma cells. Cancer Cell Int 2019;19:349. [Crossref] [PubMed]
- Lv R, Zhang QW. The long noncoding RNA FTH1P3 promotes the proliferation and metastasis of cervical cancer through microRNA-145. Oncol Rep 2020;43:31-40. [PubMed]
- Yang C, Shen S, Zheng X, et al. Long noncoding RNA HAGLR acts as a microRNA-143-5p sponge to regulate epithelial-mesenchymal transition and metastatic potential in esophageal cancer by regulating LAMP3. FASEB J 2019;33:10490-504. [Crossref] [PubMed]
- Guo X, Chen Z, Zhao L, et al. Long non-coding RNA-HAGLR suppressed tumor growth of lung adenocarcinoma through epigenetically silencing E2F1. Exp Cell Res 2019;382:111461. [Crossref] [PubMed]
- Qu Y, Zheng S, Kang M, et al. Knockdown of long non-coding RNA HOXD-AS1 inhibits the progression of osteosarcoma. Biomed Pharmacother 2018;98:899-906. [Crossref] [PubMed]
- Lu C, Ma J, Cai D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumour Biol 2017;39:1010428317697574. [Crossref] [PubMed]
- Huang JZ, Chen M. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 2017;68:171-84.e6. [Crossref] [PubMed]
- Mo JS, Park YR, Chae SC. MicroRNA 196B regulates HOXA5, HOXB6 and GLTP expression levels in colorectal cancer cells. Pathol Oncol Res 2019;25:953-9. [Crossref] [PubMed]
- Miller KR, Patel JN, Zhang Q, et al. HOXA4/HOXB3 gene expression signature as a biomarker of recurrence in patients with high-grade serous ovarian cancer following primary cytoreductive surgery and first-line adjuvant chemotherapy. Gynecol Oncol 2018;149:155-62. [Crossref] [PubMed]
- Fu H, Fu L, Xie C, et al. miR-375 inhibits cancer stem cell phenotype and tamoxifen resistance by degrading HOXB3 in human ER-positive breast cancer. Oncol Rep 2017;37:1093-9. [Crossref] [PubMed]
- Cui M, Chen M, Shen Z, et al. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J Cell Biochem 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Li Z, Rock JB, Roth R, et al. Dual stain with SATB2 and CK20/villin is useful to distinguish colorectal carcinomas from other tumors. Am J Clin Pathol 2018;149:241-6. [Crossref] [PubMed]
- Ma C, Lowenthal BM, Pai RK. SATB2 is superior to CDX2 in distinguishing signet ring cell carcinoma of the upper gastrointestinal tract and lower gastrointestinal tract. Am J Surg Pathol 2018;42:1715-22. [Crossref] [PubMed]
- Xu M, Xu X, Pan B, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer 2019;18:135. [Crossref] [PubMed]
- Yahagi M, Okabayashi K, Hasegawa H, et al. The worse prognosis of right-sided compared with left-sided colon cancers: a systematic review and meta-analysis. J Gastrointest Surg 2016;20:648-55. [Crossref] [PubMed]


