
Developing prognostic gene panel of survival time in lung adenocarcinoma patients using machine learning
Introduction
According to annual statistics reported from the American Cancer Society (1), more than 1 out of every 4 cancer deaths are due to lung cancer. About 80% of lung cancer cases are non-small cell lung cancer (NSCLC). It is classified into three pathological subtypes: adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. Lung adenocarcinoma (LUAD) is most common in young women and Asian populations, and it is associated with the mutation of some molecular targets, such as BRAF and HER2 (2).
The current treatment of NSCLC is gradually evolving from chemotherapy or radiotherapy to targeted drug therapies based on the genetic alterations, such as Osimertinib (3). Recent studies (4) found that co-occurring mutations in STK11 and TP53 in KRAS-mutant lung adenocarcinoma had an impact on tumor cell proliferation and immune surveillance responses. Another study (5) indicated that receptor-interacting serine/threonine protein kinase 4 (RIP4) is a regulator of tumor differentiation in lung adenocarcinoma. It can be inferred that genetic alterations are greatly associated with the development of NSCLC. Although previous scholars have obtained a lot of data from the microarray technique and the Next Generation Sequencing (NGS), information from these data may not be explored entirely. In this study, it is hypothesized that genetic features selected from these data will correlate with survival time of patients, which could be considered as one of the best indicators of survival and severity of illness.
During cancer treatment, doctors and patients pay close attention to survival time. Traditional survival prediction depends on the clinicopathological characteristics of patients, which is imprecise sometimes. In order to be more accurate, it is better to apply artificial intelligence to the medical domain (6,7). Cox regression model is a traditional method to predict the overall survival time of patients, but does not achieve better performance (C-indexaverage=0.58) (8). In our study, we compared eight machine learning models based on The Cancer Genome Atlas (TCGA) dataset, including DNA sequence, RNA expression and DNA methylation. We identified the correlation between genes and survival time. Then, the algorithms and the selected genes were validated using the GEO dataset, and data of DNA methylation and DNA mutation are used to further analyze the mechanism of RNA expression.
Methods
Source of data
We obtained the LUAD related data set from the TCGA portal (https://portal.gdc.cancer.gov/). For subsequent validation and analysis, we acquired the GEO dataset (GSE 72094), DNA methylation dataset and DNA mutation dataset from the GEO website (https://www.ncbi.nlm.nih.gov/gds) and the Firebrowse website (http://www.firebrowse.org/). All filtered samples (TCGA Dataset and GEO Dataset) must include the RNA-seq file, vital status and days to last follow-up.
TCGA dataset (RNA-sequence, DNA methylation, DNA mutation)
A total of 291 RNA sequencing (RNA-Seq) files, including all open source RNA sequencing data and the corresponding clinical information files, were acquired. The downloaded data was integrated and spliced with clinical data using R(v3.4.3). We merged the related information using python packages (Pandas v0.23.0 and Numpy v1.14.3) (9,10). The RNA-Seq samples were removed if they did not have the corresponding clinical files (11). The genetic data features were removed if having zero values in more than 85% patients (the number of genetic features were reduced from 60,038 to 40,540). Then, we normalized all genetic feature columns by dividing the maximum value of the column (12). And we deleted some samples based on the following reasons: (I) removed samples who were still alive but had less than two years of cancer (because we are not sure how long these samples will survive in the outcome events); (II) remove non-primary tumor samples (Figure 1).
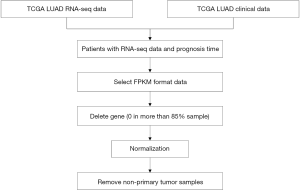
DNA methylation data is divided into two parts according to the methylation chip. One part of the samples is measured using the Illumine Human Methylation 27 Beadchip and another part using in the Illumine Human Methylation 450 Beadchip. The Methylation 27 dataset has 200 samples. For the two datasets, the samples which did not have paired RNA expression data in the TCGA were removed. One issue to note is that there are conversion problems that some genes do not have corresponding methylation probes. Thirty percent genes in the TCGA dataset cannot match the corresponding methylation probes in the methylation dataset. The limma package was used in R language to analyze the methylation data (13).
The DNA mutation level 3 dataset was downloaded from Firebrowse. We got the mutation data of 131 samples that appeared in the TCGA data set.
GSE 72094 dataset (RNA-Seq)
We processed GEO dataset in the same process of the TCGA dataset. And finally there was 174 samples in the available GEO dataset.
Feature selection
As reported in previous research, using all genes whose expression levels are measured to predict outcomes did not get a high accuracy. And 60,038 genes are first derived from RNA-Seq, and then 19,498 genes are deleted since they have zero value in 85% samples or more. In this study, we used the Relief (Relevant Features) algorithm which was first proposed by Kira and Rendell on the basis of the instance-based learning (14). We randomly selected a sample R from the training set D, then found k nearest neighbor samples H from samples of the same type as R, fiund k nearest neighbor samples M from samples of different types from R, and finally updated the feature weight according to the formula defined as follows:
where A=1,2...N, N is the number of features, m is the number of algorithm iterations. It calculates a feature score for each feature that can then be applied to rank and select top scoring features for feature selection. Using this method, we chose the top 200 features which we used for the downstream modeling. We used the first 1 to 200 features to train the model, and finally determined 22 genes based on the model accuracy (Figure 2). See below for a detailed description.
For survival prediction, patients in the test set were classified into >3 years of survival time and <3 years. The variance selection method and the chi-square test method were also utilized to select the features (not shown), and the relief algorithm was selected by comparing the outcomes.
Machine-learning algorithms for prediction
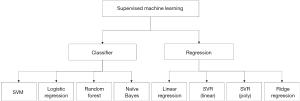
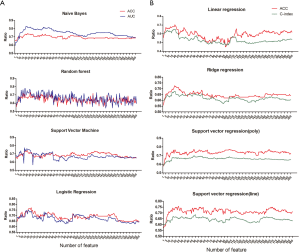
In this study, the classification methods (Figure 3) applied were Support Vector Machine (SVM) (15), Random Forest (RF) (16), Logistic Regression (LR) and Naïve Bayes (NB) (17). The regression methods applied (Figure 3) were Linear Regression, Support Vector Regression (kernel Poly), Support Vector Regression (kernel Linear), and Ridge Regression. Based on the fitting results, we could classify the samples into two categories: shorter prognosis time group (less than 3 years) and longer prognosis time group (more than 3 years). We iteratively used genes which ranked 1 to 200 to train these eight machine learning models, recorded the accuracy of each model and plotted the accuracy curve. We used 4-fold cross-validation to avoid the overfitting problems. And for the accuracy, AUC, c-index and other evaluation indicators, we calculated the average values as the final results. By comparing the accuracy curves of the eight models, we selected the optimal model and the corresponding number of features required for the model. Then predictive model was built with the selected parameters. We used this model combined with selected genes to verify on the Gene Expression Omnibus (GEO) datasets (GSE72094). We randomly set aside 20% of the total data as a test set. And the Kaplan-Meier plot were drawn (18).
Evaluation
For classification, we used accuracy and Area Under Curve (AUC) (19) to judge model quality. For the fitting, in addition to the accuracy, we used the concordance index (C-index) to evaluate the pros and cons of the fitting results. The accuracy (ACC) was the ratio of the number of correctly classified samples to the number of all samples. C-index can be seen as the fraction of all pairs of individuals whose predicted survival times are correctly ordered and is based on Harrell C statistics (20). A C-index score around 0.70 means a good model, whereas a score around 0.50 means that the fitting result represents a random guess. Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE) are indicators to measure the accuracy of regression algorithms. The smaller the value, the more accurate the algorithm.
Gene functional analysis
Gene Ontology analysis and GAD database are used in DAVID website (https://david.ncifcrf.gov/). GO is a formidable resource to understand the meaning of genes and interpret these genes. GAD database interprets the relationship of genes and diseases.
Statistical analysis
Breslow test, Mann-Whitney U test, Student’s t test and Cox model were used to analyze data in this study. Breslow test is used in survival analysis. Mann-Whitney U test is used in comparing two groups without making the assumption that values are normally distributed. Student’s t test is used in comparing the expression of different groups. Cox model is used in filtering features in the supplementary appendix. Graph Pad Prism (V5.01) and SPSS (V23) were used in statistical analysis.
Results
Survival prediction and outcome
From the TCGA-LUAD datasets, we used 131 cancer samples that covered RNA-Seq, DNA-Seq and DNA methylation. RNA-Seq was used in training and validating the predication models. DNA-Seq and DNA methylation were utilized to subsequently analyze the gene expression and discuss the relationship. We chose the 3-year survival time to divide the patient into two group and train the model. Although 5-year survival rate is the most common indicator, we would face the problem of data imbalance if we choose 5-year survival time for grouping. We preprocessed the data as described in the previous “Methods”. We used eight algorithms to predict that the patients in the validation set would survive for more than 3 years or less (Table 1).
Table 1
| Model | ACC | C-index | MAE | RMSE | AUC | Minimum number of features |
|---|---|---|---|---|---|---|
| Linear regression | 0.70 (0.57–0.84) | 0.65 (0.60–0.73) | 2.14 (1.79–2.45) | 3.01 (2.24–3.74) | – | 19 |
| Ridge regression | 0.73 (0.66–0.81) | 0.68 (0.60–0.74) | 1.91 (1.70–2.23) | 2.70 (2.24–3.74) | – | 19 |
| Line SVR | 0.75 (0.69–0.84) | 0.65 (0.53–0.74) | 1.92 (1.48–2.37) | 2.78 (2.13–3.50) | – | 24 |
| Poly SVR | 0.77 (0.69–0.81) | 0.69 (0.65–0.72) | 1.92 (1.64–2.15) | 2.81 (2.25–3.49) | – | 49 |
| Naïve Bayes | 0.75 (0.68–0.81) | – | – | – | 0.81 (0.70–0.94) | 22 |
| SVM | 0.74 (0.69–0.81) | – | – | – | 0.73 (0.62–0.81) | 16 |
| Random forest | 0.75 (0.72–0.78) | – | – | – | 0.76 (0.69–0.83) | 55 |
| Logistic regression | 0.77 (0.68–0.84) | – | – | – | 0.74 (0.56–0.93) | 24 |
We filtered the methods of the accuracy >75% with the corresponding number of features <25-Naïve Bayes, SVR (line) and logistic regression- applying to the following confirmation cohort. ACC, accuracy; AUC, area under curve; MAE, mean absolute error; RMSE, root mean squared error; SVM, support vector machines; SVR, support vector regression.
(I) As for regression, the Poly SVR has the best performance of 77% (69–81%) while C-index is 0.69 (0.65–0.72) showing great prediction. The Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE) also have low scores of 1.92 and 2.81, respectively. (II) With respect to classification, the prediction results are shown in Table 1. Logistic regression outperforms three other algorithms with a predictive accuracy of 77% (68–84%), compared to 75% (70–94%) accuracy of Naïve Bayes. C-index, MAE and RMSE are suitable for fitting methods rather than the classification, so AUC is used to evaluate the performance of the classification model. The AUC of the Logistic regression and the Naïve Bayes are 0.74 (0.56–0.93) and 0.81 (0.70–0.94) respectively (Table 1). Figure S1 shows the outcomes of accuracy and other index as the number of features increases. These results demonstrate that our methods of selecting genomic features is effective, and the predication algorithms is robust to predict survival time. Excessive genetic features used in the algorithm are considered to have the tendency of overfitting. Therefore, less genetic features and 4-fold cross validation experiment are used to avoid the overfitting problem. Naïve Bayes, the SVR (line) and logistic regression are chosen to predict survival on confirmation cohorts because they have accuracy >75% and corresponding features <25.
Validation of the prediction model
To confirm the robustness of our models, we tried to validate our models on another cohort (GSE 72094, n=174). The GSE 72094 calculate the logarithmic value of a provided number of base 2 and IRON normalized signal while TCGA data is FPKM type and the format of data preprocessing is different from TCGA. Therefore, the weights of genetic features were trained again and corresponding features is same. The training set and test set for each method were randomly selected by R package (the random library). Due to the limitation of the dataset, the result we presented in the validation set is a floor outcome of the prediction model.
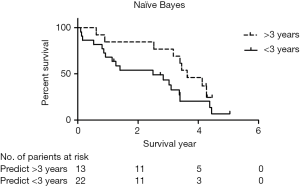
We used SVR (line), Naïve Bayes, and Logistic Regression to predict survival on confirmation cohorts. The accuracy results of the GEO dataset are as follows: SVR (poly) 57%, Naïve Bayes 69%, and Logistic Regression 51%. Logistic Regression and SVR (poly) have the highest accuracy on the TCGA cohorts, as they have low accuracy on GEO cohorts, suggesting that they are unstable on different datasets. So, Naïve Bayes is the best and the most stable algorithm for predicting survival time in lung adenocarcinoma. Naïve Bayes can significantly distinguish two cohorts (Figure 4, P=0.0438, Breslow Test).
The deep mining of the genomic features
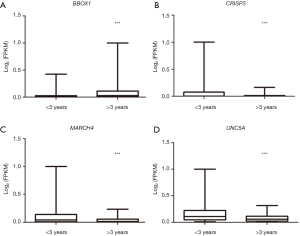
The RNA expression of 22 genes have differences in the two groups with a prognosis of less than 3 years or more. The expression of some genes is implicated in survival time of LUAD (Figure 5).
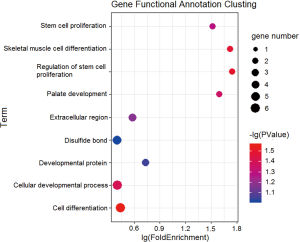
The 22 genomic features (Figure 2) include the expression data of 13 coding DNA and 9 long non-coding RNA (lncRNA), suggesting that lncRNA dose affect protein coding (21). Association between the selected genes and lung illness or cancer are shown in the GO and GAD analysis (Figure S2). We attempted to explain the change of RNA expression and find whether transcription present the association between DNA mutation and DNA methylation. Therefore, the coupled DNA-seq and DNA methylation files were downloaded from Firebrowse, where the data is acquired from the TCGA.
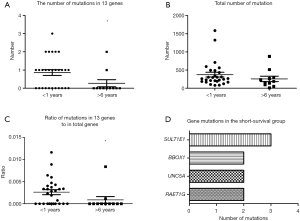
In the DNA mutation data, the mutation counts in the long-survival group (>6 years) is less than the short-survival group (<1 year) in 22 genes (Figure 6A, P=0.031, Mann-Whitney U test). Considering whether the total mutation burden in the long-survival group is less than in the short-survival group, we compared the two groups using Mann-Whitney U test (Figure 6B, P=0.147). For the ratio of mutations in 22 genes to in total genes, the short-survival group is higher than the long-survival group (Figure 6C, P=0.026, Mann-Whitney U test). Within the range of 22 genes, many mutation sites occur in the 28 samples of short-survival group, with up to 3 in SULT1E1 gene. The following mutations are BBOX1, UNC5A, RAET1G (Figure 6D). Those mutations including reported and unreported ones may be associated with cancer progression affecting the survival of patients. Epigenetic alterations are reported to play an essential role in the transcription of gene, as we know. However, we did not find the significant correlation between the DNA methylation value and survival time using the limma package applying the filter of |log2FC|>1 and FDR <0.05 (13).
Discussion
Transcriptome data analysis captures coding and non-coding genes and quantifies the difference of gene expression in cells, tissues and organs (22). The knowledge of genes has influenced our clinical treatments of illnesses, even our lifestyle. Some researchers have attempted to associate the clinical features, medical images or living habits with the survival of lung cancer patients, but the outcomes were not of great prognostic value. In this study, we used the transcription files of two distinct groups (<3 years group and >3 years group) to train the feature filtering algorithm to acquire the weights of genetic features, and then predicted whether the patients’ survival time is <3 years or >3 years.
To manifest the robustness of our survival model, we applied this algorithm on the GEO dataset and achieved a consistent performance on the GEO confirmation cohort. In the confirmation cohorts, accuracy of outcomes dropped from 75% to 69%. The reasons may be as follows: Firstly, the GEO dataset used the microarray methods to calculate the value of RNA expression, resulting that gene features in the GEO dataset do not include lncRNA and micro RNA. Secondly, the format of data processing in GEO dataset is different from the TCGA LUAD dataset. In general, the results in the GEO dataset can confirm the feasibility of the algorithm. Because of the instability of Logistic Regression and SVR(poly), Naïve Bayes shows the best performance in prediction.
In some significant genes, some gene expression is associated with the survival time (Figure 3). Although other genes do not show significant differences, we cannot rule out the possibility that genes are not differently expressed and trace amounts of protein may affect the biological function. In our research, 22 genetic features are selected from Relief and Naïve Bayes. Some genetic features have been reported in previous research. The UNC5A feature is the top significant gene with the highest weight. UNC5A is down-regulated in multiple tumors including lung cancer, may be tumor suppressor inhibiting tumor extension (23). Low expression in CRISP3 predict a good prognosis in breast cancer (24). ANXA13 is up-regulated in colorectal cancer and may be associated with metastasis (25). SOX11 contributes to increase invasive growth and the progression of ductal carcinoma in situ to invasive breast cancer (26). SULT1E1 could suppress tumor proliferation and invasion in mammary cancer model (27). The filtered features we screened are biologically significant and are worthwhile to explore. However, the expression of MARCH4, RAET1G, PAMR1 and other genes are not reported in the field of lung cancer.
As shown in the Figure 3, there is a negative correlation between the number of mutations in 22 genes and survival time. The mutation number in 22 genes, rather than the total mutation number, is greater in the short-survival cohort. It indirectly confirms that 22 genetic features affect the survival time of LUAD indeed. But we did not find any differences regarding gene methylation between the two groups (<3 years and >3 years groups).
There are some limitations on combining the machine learning and RNA expression. For instance, the algorithms can only process data, not the potential relationship between the data. MARCH4, RAET1G, PAMR1 are on the 22-gene panel of predicting the survival time, and we do not know whether these three genes are incorrectly associated on the list of 22 genes or if they have just not been reported yet. In addition, transcriptome data is characterized by a small sample size but a large number of features, limiting many deep learning algorithms which are suitable.
Conclusions
In conclusion, we found that there is correlation between the expression of some genes and survival time. The model of 22-gene panel could predict survival time of lung adenocarcinoma patients by Naïve Bayes algorithm. Using this approach, we filtered some specific genes, and this would be helpful for doctors’ diagnosis and patients’ treatment.
Acknowledgments
Thanks for William Donelan in University of Florida to review and modify the manuscript.
Funding: This work was funded by
Availability of Data and Material: The code during the current study is available in https://github.com/ningshuishi/genedata.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2739). MY, WW and DT report grants from National Natural Science Foundation of China, grants from the Major Science and Technology Innovation Project of Shandong Province, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This article does not contain any studies with human participants or animals performed by any of the authors.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Calvayrac O, Pradines A, Pons E. Molecular biomarkers for lung adenocarcinoma. Eur Respir J 2017;49:1601734. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-125. [Crossref] [PubMed]
- Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209-16. [Crossref] [PubMed]
- Kopparam J, Chiffelle J, Angelino P, et al. RIP4 inhibits STAT3 signaling to sustain lung adenocarcinoma differentiation. Cell Death Differ 2017;24:1761-71. [Crossref] [PubMed]
- Deo RC. Machine Learning in Medicine. Circulation 2015;132:1920-30. [Crossref] [PubMed]
- Zhang L, Tan J, Han D, et al. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today 2017;22:1680-5. [Crossref] [PubMed]
- Liu C, Wang X, Genchev GZ, et al. Multi-omics facilitated variable selection in Cox-regression model for cancer prognosis prediction. Methods 2017;124:100-107. [Crossref] [PubMed]
- Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model 1999;17:57-61. [PubMed]
- Demsar J, Curk T, Erjavec A, et al. Orange: data mining toolbox in Python. J Mach Learn Res 2013;14:2349-53.
- Title of subordinate document. In: mRNA Analysis Pipeline. Available online: https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/Expression_mRNA_Pipeline/. Accessed April, 2019.
- Allison DB, Cui X, Page GP, et al. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 2006;7:55. [Crossref] [PubMed]
- Yang IV, Pedersen BS, Rabinovich E, et al. Relationship of DNA Methylation and Gene Expression in Idiopathic Pulmonary Fibrosis (IPF). Am J Respir Crit Care Med 2014;190:1263-72. [Crossref] [PubMed]
- Kira K, Rendell L. The feature selection problem: traditional methods and a new algorithm. In: Proceedings of the national conference on artificial intelligence. John Wiley & Sons Ltd, 1992;129.
- Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273-29. [Crossref]
- Ho TK. The random subspace method for constructing decision forests. IEEE Trans Pattern Anal Mach Intell 1998;20:832-44. [Crossref]
- Friedman N, Sheng S. Bayesian Network Classifiers. Machine Learning 1997;29:131-63. [Crossref]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958;53:457-81. [Crossref]
- Bamber D. The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J Math Psychol 1975;12:387-415. [Crossref]
- Uno H, Cai T, Pencina MJ, et al. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 2011;30:1105-17. [PubMed]
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484-8. [Crossref] [PubMed]
- Jiang Z, Zhou X, Li R, et al. Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol Life Sci 2015;72:3425-39. [Crossref] [PubMed]
- Thiebault K, Mazelin L, Pays L, et al. The netrin-1 receptors UNC5H are putative tumor suppressors controlling cell death commitment. Proc Natl Acad Sci U S A 2003;100:4173-8. [Crossref] [PubMed]
- Wang Y, Sheng N, Xie Y, et al. Low expression of CRISP3 predicts a favorable prognosis in patients with mammary carcinoma. J Cell Physiol 2019;234:13629-38. [Crossref] [PubMed]
- Jiang G, Wang P, Wang W, et al. Annexin A13 promotes tumor cell invasion in vitro and is associated with metastasis in human colorectal cancer. Oncotarget 2017;8:21663-73. [Crossref] [PubMed]
- Oliemuller E, Kogata N, Bland P, et al. SOX11 promotes invasive growth and ductal carcinoma in situ progression. J Pathol 2017;243:193-207. [Crossref] [PubMed]
- Xu Y, Lin X, Xu J, et al. SULT1E1 inhibits cell proliferation and invasion by activating PPARγ in breast cancer. J Cancer 2018;9:1078-87. [Crossref] [PubMed]



