Knockdown of ferritin heavy chain (FTH) inhibits the migration of prostate cancer through reducing S100A4, S100A2, and S100P expression
Introduction
Prostate cancer (PCa) is the deadliest cancer after lung cancer in men and also has the highest incidence of cancer worldwide (1). Tissue biopsy is the gold standard for the diagnosis of the prostate (2), while serum prostate-specific antigen (PSA) remains the most widely used biomarker for the management of early PCa (3). Although PSA has extensive clinical applications, increased PSA levels can be found in benign prostatic hyperplasia (BPH) and other diseases (4,5). When the patient’s total prostate-specific antigen (tPSA) level is in the gray area (4–10 ng/mL), the diagnostic specificity is only 25–40% and may cause unnecessary biopsy in some patients (3,5). Therefore, the development of more sensitive and specific tumor markers and oncogenes to improve the diagnostic accuracy and prognosis in PCa is urgently needed.
Serum ferritin, the storage form of iron, is elevated in many diseases and has emerged as a novel biomarker for many kinds of cancers (6-10). With a growing number of studies indicating that ferritin plays a vital role in oncogenesis, researchers have attempted to use ferritin as a biomarker for disease detection, therapy monitoring, and drug resistance surveillance (9,11-14). According to Wang et al., serum ferritin—binding PSA detection can improve the diagnostic accuracy of PCa (12). In PCa patients, serum ferritin is significantly correlated with total PSA (tPSA), free PSA (fPSA), and fPSA-tPSA ratio, withslight correlations found in the corresponding BPH controls (15). However, the role of ferritin in the progress of PCa is still unclear. Studying the mechanism of ferritin in the progression of PCa is of great significance for early diagnosis, treatment, and prognosis.
The S100 protein family, which comprises 21 members, has a high degree of structural similarity. Nonetheless, the members are functionally non-interchangeable (16). This protein family regulates cellular responses through the function of intracellular Ca2+ sensors and extracellular factors. An increasing number of studies have shown that dysregulation of S100 protein expression is a common event in many human cancers (17). Elevated levels of S100P were found to be correlated with progression to metastatic disease. The disorder of S100P is involved in multiple types of tumorigenesis (18-21). Meanwhile, S100A4 has been implicated in invasion and metastasis, and upregulation of its expression can be seen in a variety of tumor cells, including in prostate, pancreatic, breast, ovarian, renal, and brain tumors (22-28). Also, S100A2 was dysregulated in prostate and breast cancer and found to have tumor-suppressing effects (29,30). Moreover, other proteins, like S100A6 and S100A10 are dysregulated in multiple kinds of cancers (31-33).
Our earlier studies have found that urinary protein has potential application value in the diagnosis of PCa. In particular, ferritin may be useful as a marker for the differential diagnosis of PCa and BPH (13), and the decreased expression of ferritin heavy chain (FTH) was found to significantly reduce the migration and proliferation of human prostate cancer cells (PC3) (34). Based on our earlier study and the role of FTH in PCa, we looked to explore the mechanisms of FTH in tumor cell migration.
To identify the downstream target gene of FTH in the pathogenesis of PCa, isobaric tags for relative and absolute quantitation (iTRAQ) tandem mass spectrometry was performed to analyze the changes of PC3 cell protein spectrum after FTH gene knockdown. The results of mass spectrometry showed that compared with the normal control (NC) group, 420 proteins were downregulated while 442 proteins were elevated in FTH silenced PC3 cells that generated by small hairpin (sh) RNA lentiviral transfection (LV-shFTH). The expression of protein S100A4, protein S100A2, protein S100P, which are migration-associated proteins considered to be involved in cell migration and invasion (18,19,22,25), were reduced. Therefore, we hypothesized that the low expression of FTH in PC3 cells caused the low expression of S100A4, S100A2, and S100P. Since these migration-related genes are suppressed, the process of epithelial-mesenchymal transition (EMT) transformation would be weakened, subsequently causing a decrease in cell migration. Our experiments demonstrate that decreased expression of FTH can induce decreased expression of S100A4, S100A2, and S100P, and may provide a theoretical basis for FTH as a new tumor marker for the early diagnosis of PCa.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-19-2852).
Methods
Cell selection and culture
Androgen-independent prostate cancer epithelial cells (PC3), which are the androgen-independent PCa cells isolated from human bone metastatic PCa tumor tissues, were purchased from the National Infrastructure of Cell Line Resource (CRL1435, Beijing, China). Stable FTH-silenced cell lines were generated by LV-shFTH, while the negative control group was only infected with empty virus (NC); both types of cells had already been established in our laboratory (34). Cells were cultured in F-12 Kaighn Modification (Gibco) supplemented with 10% fetal bovine serum (Zhejiang Tianhang Biotechnology Co. Ltd., Zhejiang, China), and grown in incubators with 5% CO2 at 37 °C. They were digested with 0.05% trypsin (Gibco) for subculturing.
Protein preparation
An appropriate amount of lysis buffer was added (7M urea, 2M thiourea, 0.1% CHAPS) to the cell sample, with vortex mixing (with added protease inhibitor and the ratio of lysate and protease inhibitor is 50:1), sonication for 60 s, at 30% amplitude. After ultrasonication, the sample was centrifuged at 12,000 g for 15 min. The supernatant was extracted, dispensed, with 10 uL being quantified, and the rest being frozen at −80 °C. The total protein concentration was measured by the Bradford protein assay.
iTRAQ analysis of protein expression changes in FTH low expression PC3 cells
iTRAQ was performed for proteomics measurements in both LV-shFTH PC3 cells and NC group cells. The experiment was repeated 3 times for each sample in this experiment. Data obtained by mass was processed using the UniProt Homo sapiens database (https://www.uniprot.org/). The mass spectrometry analysis of iTRAQ was completed by Thermo Q Exactive mass spectrometry. The original mass spectrometry documents were processed by Proteome Discoverer 1.4, a commercial software manufactured by Thermo Fisher Scientific (retrieval parameter settings are shown in Table 1). P values <0.05 and fold change >1.2 were the standards set to screen the target protein.
Table 1
| Parameter | Value |
|---|---|
| Enzyme | Trypsin |
| Static modification | C carboxyamidomethylation (57.021 Da) |
| Dynamic modification | M Oxidation (15.995 Da); N-terminal, K, iTRAQ 8 plex |
| Species | Homo sapiens |
| Precursor ion mass tolerance | ±15 ppm |
| Fragment ion mass tolerance | ±20 mmu |
| Max missed cleavages | 2 |
Western blot
Cellular proteins were extracted using radio immunoprecipitation assay (RIPA) lysis buffer, and protease inhibitors were added to the lysate to protect the proteins. Anti-FTH polyclonal antibody (ab65080, Abcam), anti-αS1002/S100A2 (ab109494, Abcam), S100A4 (ab124805, Abcam), and S100P (ab133554, Abcam) monoclonal antibodies were diluted as the first antibody in a 1:1,000 ratio. Anti-mouse IgG H&L horseradish peroxidase (HRP), goat anti-rabbit IgG H&L HRP, and mouse anti-β actin monoclonal antibodies (Beijing Dingguo Chang Sheng Company) were diluted to a 1:2,500 ratio. Primary antibodies and polyvinylidene fluoride (PVDF) membranes were incubated overnight at 4 °C. The next day, the PVDF membranes were washed 3 times in phosphate-buffered saline (PBS) buffer. Then, they were incubated with secondary antibodies, which were labeled with HRP for 1 h. Target protein bands were determined by enhanced chemiluminescence, and β-actin was an internal reference to achieve consistent sample loading per well. Each experiment was repeated 3 times.
Statistical analysis
We use the mean ± standard deviation (SD) to stand for the data. All data were collected from independent experiments. Experimental results were statistically analyzed using GraphPad Prism 6.0 statistical software (GraphPad Software Inc., La Jolla, CA, USA). SPSS 17.0 software (IBM Corp., Armonk, NY, USA) was used to detect the differences between the 2 cell groups, which were assessed using Student’s t-test. P values <0.05 were considered statistically significant results.
Compliance with ethical standards
Statement of ethics approval
The study did not require ethical approval as it did not contain any research on human participants or animals. The authors declare that they have no competing interests.
Funding
This work was supported by the Enhancement Funding of Beijing Key Laboratory of Urinary Cellular Molecular Diagnostics (no. 2020-JS02).
Results
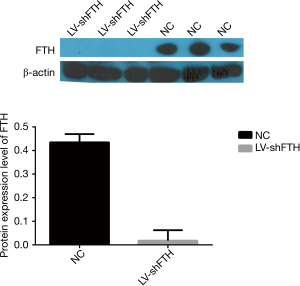
Western blot assay to confirm the effect of FTH-silencing
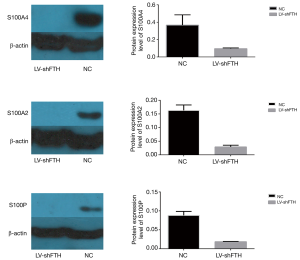
The cells used in this experiment were FTH-silencing PC3 cell lines previously established in our laboratory. Therefore, the FTH expression of cells was confirmed by Western blot before the later experiments (Figure 1). A low level of FTH expression was seen in the LV-shFTH PC3 cells as compared with the cells of the NC group.
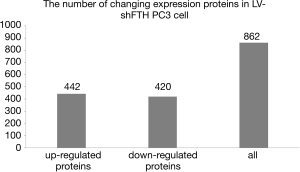
iTRAQ mass spectrometry analysis results
iTRAQ mass spectrometry analysis was used to quantify changes in protein expression after FTH knockdown in PC3 cells. The data with P≤0.05 and difference ratio ≥1.2 were selected for further analysis. Compared with the NC group, the expression of 420 proteins was downregulated, while the expression of 442 proteins was elevated in LV-shFTH PC3 cells, according to mass spectrometry (Figure 2).
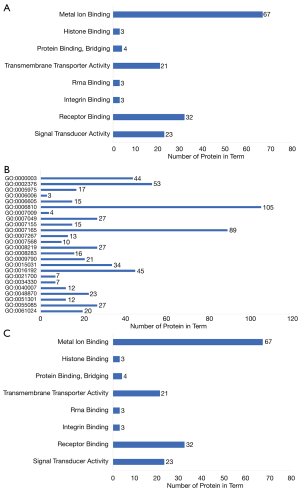
Analysis of the downregulated expression of genes: Gene Ontology (GO) enrichment
The downregulated proteins were mapped to each term of the GO database. The result of GO enrichment analysis is shown in Figure 3; the molecular function (MF) of the changing genes is shown in Figure 3A; the biological processes (BP) in which the differential genes were involved in are shown in Figure 3B and the location of the changing proteins are shown in Figure 3C. The items shown in the figures indicate a significance level of less than 0.05 by Fisher’s exact test in each category. According to the GO enrichment analysis, metal ion binding and receptor binding were the top 2 molecular functions most affected in the changing proteins (Figure 3A). The BP of the differential proteins were included in transport and signal transduction, among other processes (Figure 3B), and the cellular component of the differential proteins was mainly involved in the cytoplasm and membrane (Figure 3C). These targets are the first screening results of mass spectrometry and have not been verified by other experiments.
FTH knockdown was accompanied by the significantly downregulated expression of S100A4, S100A2, and S100P
To further narrow the scope of research, the downregulated proteins were selected for in-depth analysis. It was found that the expression of tumor cell migration-related genes, S100A4, S100P, and S100A2, were reduced significantly (Table 2). Of all the differentially expressed proteins, the changing degree of S100A4, S100A2, and S100P ranked first, second, and fourth, respectively. The spectrum peptide fingerprint results of these 3 proteins correspond to Figures 4-6, respectively. The expression level of S100A4, S100A2, and S100P was then confirmed by Western blot. As shown in Figure 7, S100A4, S100P, and S100A2 were significantly reduced after FTH knockdown in PC3 cell lines. These results show that low FTH expression affected the activity of S100 proteins.
Table 2
| Gene name | Protein name | Ratio | P value |
|---|---|---|---|
| S100A4 | S100 calcium-binding protein A4 (Metastasin) | 0.29 | <0.05 |
| S100A2 | S100 calcium-binding protein A2 | 0.39 | <0.05 |
| S100P | S100 calcium-binding protein P (Migration-inducing gene 9 protein) | 0.45 | <0.05 |
Discussion
The early diagnosis and evaluation of the stage of progression for prostate cancer are crucial for the treatment and prognosis of PCa patients. A large number of researchers are devoted to finding more sensitive and specific tumor markers for PCa as a supplement to PSA detection (5,12,35). Recent research has highlighted the fact that iron metabolism abnormalities exist in prostate cancer and that inadequate ferritin acquisition slows down the growth and invasiveness of cancer cells. Plenty of studies have been conducted to elucidate how ferritin affects the development of PCa (9,34,36). For instance, iron-responsive element-binding proteins (IRPs) were critical for supporting intracellular iron homeostasis. Researchers also found that the upregulation of IRP2 in concert with normal iron regulatory mechanisms could facilitate iron retention and promote tumor growth in prostate cancer cells (36). Furthermore, it was found that nuclear transcription factor Nrf2 could suppress prostate cancer cell viability, migration, and mitosis through upregulating ferroportin (11). However, despite these findings, we still have limited knowledge of the molecular mechanisms behind ferritin regulation in PCa.
Preliminary analysis of the changing protein expression in PC3 cells after knocking down FTH by mass spectrometry and subsequent Western blot analysis further showed that S100A4, S100A2, and S100P proteins were significantly reduced in LV-shFTH PC3 cells compared to NC. These results suggest that the low expression level of FTH can affect S100 family protein expression. A large number of studies report that S100 proteins are closely related to tumor cell migration (19,22,25,37). Many S100 family proteins are involved in cell migration/invasion and regulating cytoskeletal dynamics, including epithelial-mesenchymal transition (EMT), which is a typical process and involves S100A2, S100A4, S100A6, S100A7, S100A14, and S100P (38). Researchers have found that S100A4 is positively correlated with Zinc Finger E box–binding protein-1 (ZEB1) and suppressed by E-cadherin. ZEB1 protein takes part in embryonic development and formation and is a member of the zinc finger transcription factor family. Coincidentally, ZEB1 can also influence E-cadherin expression. Moreover, another study found that the use of small interfering RNAs reduced the expression of ZEB1 and subsequently enhanced tumor cell migration and invasive and proliferative capacity (23). These results inspired us to use iTRAQ analysis, which revealed that ZEB1 is downregulated in LV-shFTH PC3 cells (P≤0.05, difference ratio ≥1.2), for a more specific mechanism of how FTH affect S100 Protein. Based on the investigations above, a reasonable assumption can be made that FTH knockdown weakens cell migration through 3 possible pathways via the S100 protein: (I) low FTH expression directly acts on the S100 protein through certain molecular mechanisms; (II) low FTH expression first affects ZEB1, which then influences S100 protein expression; (III) both of the above approaches co-occur. We will try further to elucidate the specific mechanisms in a future study.
Members of the S100 protein family are abnormally expressed in distinct types of tumors, which usually induce their upregulation. The mechanism related to how the S100 protein family acts in tumors is complex. Studies have shown that breast cancers expressing high levels of S100A4 have a significantly worse prognosis compared to those negative for S100A4 (39). It has also been found that the overexpression of S100A4 may be enough to induce the development of metastatic phenotypes in human breast tumor cells. Similarly, S100A4 was discovered to be overexpressed during the progression of PCa not only in humans but also in the transgenic adenocarcinoma of the mouse prostate (TRAMP) mouse model (40,41). Additionally, S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9 (MMP9) (22). A study conducted by Bjornland et al. also concluded that S100A4 not only stimulates the motility of tumor cells but may also exert an effect on tumor invasion characteristics by affecting the expression of matrix metalloproteinases (MMPs) and their endogenous inhibitors, contributing to metastasis formation (42). Overwhelming evidence shows a complex relationship between the S100A4 gene product and the cytoskeleton. Investigation on how the S100A4 gene contributes to cell migration is crucial to further developing the treatment and prognosis of PCa.
Similar to S100A4, elevated levels of S100P in PCa were strongly correlated with progression to metastatic disease. Some researchers modulated S100P levels in different prostate cancer cells by silencing S100P levels in 22Rv1 cells and by overexpressing S100P in PC3 cells. Their results confirmed that overexpression of S100P could upregulate the expression of androgen receptor and promote the progression of prostate cancer by promoting cell growth, while a prominent cytostatic effect was induced by silencing S100P in 22Rv1 cells (21). Meanwhile, S100P was found to induce dissociation of nonmuscle myosin II (NMIIA) filaments leading to a weakening of focal adhesion sites (FAS), which resulted in a reduction in cell adhesion and the promotion of cell migration, both of which are critical in the metastatic cascade (43). Increased expression of androgen receptor in S100P-overexpressed cells may promote the progression of prostate cancer by accelerating cell growth. Also, S100P can be considered a potential drug target or chemosensitizing target, and can also be a biomarker for invasive, hormone-refractory, and metastatic prostate cancer (21,35,43). Several studies have explored the role of S100A2 in the tumor, and intriguingly, have found 100A2 to have tumor-suppressing effects. Buckley et al. identified S100A2 as a BRCA1/p63 coregulated tumor suppressor gene with roles in regulating mutant p53 stability (44). Another studied showed that knockdown of S100A2 expression could compromise TGF-β1-induced cell migration and invasion of Hep3B cells, thus suggesting the role of S100A2 in mediating TGF-β-induced migration/invasion of tumor cells (45).
Many studies have indicated that FTH can be a diagnostic biomarker for PCa, with some asserting that S100 family proteins are associated with tumor cell migration (in vitro) and tumor metastasis (in vivo). The findings of our research demonstrate that the expression level of FTH affects S100 family protein expression in PC3 cells (Figure 8). Low expression of FTH in PC3 cells will cause a decrease in the expression of specific proteins of the S100 protein family, which are related to cell migration. Combined with the prelaboratory study on the decline in the migration ability of FTH knockdown PC3 cells, it is reasonable to assume that proteins such as S100A4, S100A2, and S100P are involved in the migration and invasion ability weakened by FTH knockdown of PC3 cells. These findings give us a new perspective on the treatment of PCa: it may be possible to find modulators to inhibit the effects of FTH-regulated genes, such as the S100 family, and inhibiting the migration of PCa cells, thus mitigating the metastasis and spread of PCa.
Prospect
We believe that FTH might be a novel target in the therapy of primary and metastatic lesions of the prostate. We will endeavor to elucidate further the molecular regulatory mechanism behind the interaction of FTH and the S100 family members. Future experiments, such as those setting up tumor xenograft models in nude mice, will be taken into consideration. We will also seek to verify the relationship between other differentially expressed proteins and ferritin after FTH knockdown and clarify their effects on PCa cells.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-19-2852
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2852
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2852). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study did not require ethical approval as it did not contain any research on human participants or animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Litwin MS, Tan HJ. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA 2017;317:2532-42. [Crossref] [PubMed]
- Popov SV, Guseinov RG, Skryabin ON, et al. Prognostic significance of prostate-specific antigen in defining indications for initial prostate biopsy. Urologiia 2018;92-7. [Crossref] [PubMed]
- Chistiakov DA, Myasoedova VA, Grechko AV, et al. New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin Cancer Biol 2018;52:9-16. [Crossref] [PubMed]
- Nguyen-Nielsen M, Borre M. Diagnostic and Therapeutic Strategies for Prostate Cancer. Semin Nucl Med 2016;46:484-90. [Crossref] [PubMed]
- Cullis JO, Fitzsimons EJ, Griffiths WJ, et al. Investigation and management of a raised serum ferritin. Br J Haematol 2018;181:331-40. [Crossref] [PubMed]
- Lee S, Jeon H, Shim B. Prognostic Value of Ferritin-to-Hemoglobin Ratio in Patients with Advanced Non-Small-Cell Lung Cancer. J Cancer 2019;10:1717-25. [Crossref] [PubMed]
- Bertoli S, Paubelle E, Berard E, et al. Ferritin heavy/light chain (FTH1/FTL) expression, serum ferritin levels, and their functional as well as prognostic roles in acute myeloid leukemia. Eur J Haematol 2019;102:131-42. [Crossref] [PubMed]
- Quintana Pacheco DA, Sookthai D, Graf ME, et al. Iron status in relation to cancer risk and mortality: Findings from a population-based prospective study. Int J Cancer 2018;143:561-9. [Crossref] [PubMed]
- Vela D. Iron Metabolism in Prostate Cancer; From Basic Science to New Therapeutic Strategies. Front Oncol 2018;8:547. [Crossref] [PubMed]
- Xue D, Zhou C, Shi Y, et al. Nuclear transcription factor Nrf2 suppresses prostate cancer cells growth and migration through upregulating ferroportin. Oncotarget 2016;7:78804-12. [Crossref] [PubMed]
- Wang X, An P, Zeng J, et al. Serum ferritin in combination with prostate-specific antigen improves predictive accuracy for prostate cancer. Oncotarget 2017;8:17862-72. [Crossref] [PubMed]
- Su Q, Lei T, Zhang M. Association of ferritin with prostate cancer. J BUON 2017;22:766-70. [PubMed]
- Huang H, Qiu Y, Huang G, et al. Value of Ferritin Heavy Chain (FTH1) Expression in Diagnosis and Prognosis of Renal Cell Carcinoma. Med Sci Monit 2019;25:3700-15. [Crossref] [PubMed]
- Lein M, Stephan C, Jung K, et al. Relation of free PSA/total PSA in serum for differentiating between patients with prostatic cancer and benign hyperplasia of the prostate: which cutoff should be used? Cancer Invest 1998;16:45-9. [Crossref] [PubMed]
- Heizmann CW, Fritz G, Schafer BW. S100 proteins: structure, functions and pathology. Front Biosci 2002;7:d1356-68. [PubMed]
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer 2015;15:96-109. [Crossref] [PubMed]
- Kikuchi K, McNamara KM, Miki Y, et al. S100P and Ezrin promote trans-endothelial migration of triple negative breast cancer cells. Cell Oncol (Dordr) 2019;42:67-80. [Crossref] [PubMed]
- Nakayama H, Ohuchida K, Yonenaga A, et al. S100P regulates the collective invasion of pancreatic cancer cells into the lymphatic endothelial monolayer. Int J Oncol 2019;55:211-22. [Crossref] [PubMed]
- Gibadulinová A, Barathova M, Kopacek J, et al. Expression of S100P protein correlates with and contributes to the tumorigenic capacity of HeLa cervical carcinoma cells. Oncol Rep 2005;14:575-82. [Crossref] [PubMed]
- Basu GD, Azorsa DO, Kiefer JA, et al. Functional evidence implicating S100P in prostate cancer progression. Int J Cancer 2008;123:330-9. [Crossref] [PubMed]
- Saleem M, Kweon MH, Johnson JJ, et al. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc Natl Acad Sci U S A 2006;103:14825-30. [Crossref] [PubMed]
- Al-Ismaeel Q, Neal CP, Al-Mahmoodi H, et al. ZEB1 and IL-6/11-STAT3 signalling cooperate to define invasive potential of pancreatic cancer cells via differential regulation of the expression of S100 proteins. Br J Cancer 2019;121:65-75. [Crossref] [PubMed]
- Chong HI, Lee JH, Yoon MS, et al. Prognostic value of cytoplasmic expression of S100A4 protein in endometrial carcinoma. Oncol Rep 2014;31:2701-7. [Crossref] [PubMed]
- Li F, Shi J, Xu Z, et al. S100A4-MYH9 Axis Promote Migration and Invasion of Gastric Cancer Cells by Inducing TGF-beta-Mediated Epithelial-Mesenchymal Transition. J Cancer 2018;9:3839-49. [Crossref] [PubMed]
- Chow KH, Park HJ, George J, et al. S100A4 Is a Biomarker and Regulator of Glioma Stem Cells That Is Critical for Mesenchymal Transition in Glioblastoma. Cancer Res 2017;77:5360-73. [Crossref] [PubMed]
- Link T, Kuhlmann JD, Kobelt D, et al. Clinical relevance of circulating MACC1 and S100A4 transcripts for ovarian cancer. Mol Oncol 2019;13:1268-79. [Crossref] [PubMed]
- Küper C, Beck FX, Neuhofer W. NFAT5-mediated expression of S100A4 contributes to proliferation and migration of renal carcinoma cells. Front Physiol 2014;5:293. [Crossref] [PubMed]
- Hibi K, Fujitake S, Takase T, et al. Identification of S100A2 as a target of the DeltaNp63 oncogenic pathway. Clin Cancer Res 2003;9:4282-5. [PubMed]
- Wicki R, Franz C, Scholl FA, et al. Repression of the candidate tumor suppressor gene S100A2 in breast cancer is mediated by site-specific hypermethylation. Cell Calcium 1997;22:243-54. [Crossref] [PubMed]
- Rehman I, Cross SS, Azzouzi AR, et al. S100A6 (Calcyclin) is a prostate basal cell marker absent in prostate cancer and its precursors. Br J Cancer 2004;91:739-44. [Crossref] [PubMed]
- Zhang J, Zhang K, Jiang X, et al. S100A6 as a potential serum prognostic biomarker and therapeutic target in gastric cancer. Dig Dis Sci 2014;59:2136-44. [Crossref] [PubMed]
- Dudley KJ, Revill K, Whitby P, et al. Genome-wide analysis in a murine Dnmt1 knockdown model identifies epigenetically silenced genes in primary human pituitary tumors. Mol Cancer Res 2008;6:1567-74. [Crossref] [PubMed]
- Zhao H, Zhao X, Lei T, et al. Screening, identification of prostate cancer urinary biomarkers and verification of important spots. Invest New Drugs 2019;37:935-47. [Crossref] [PubMed]
- Mousses S, Bubendorf L, Wagner U, et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res 2002;62:1256-60. [PubMed]
- Deng Z, Manz DH, Torti SV, et al. Iron-responsive element-binding protein 2 plays an essential role in regulating prostate cancer cell growth. Oncotarget 2017;8:82231-43. [Crossref] [PubMed]
- Gross SR, Sin CG, Barraclough R, et al. Joining S100 proteins and migration: for better or for worse, in sickness and in health. Cell Mol Life Sci 2014;71:1551-79. [Crossref] [PubMed]
- Chen H, Xu C, Jin Q, et al. S100 protein family in human cancer. Am J Cancer Res 2014;4:89-115. [PubMed]
- de Silva Rudland S, Martin L, Roshanlall C, et al. Association of S100A4 and osteopontin with specific prognostic factors and survival of patients with minimally invasive breast cancer. Clin Cancer Res 2006;12:1192-200. [Crossref] [PubMed]
- Gupta S, Hussain T, MacLennan GT, et al. Differential expression of S100A2 and S100A4 during progression of human prostate adenocarcinoma. J Clin Oncol 2003;21:106-12. [Crossref] [PubMed]
- Saleem M, Adhami VM, Ahmad N, et al. Prognostic significance of metastasis-associated protein S100A4 (Mts1) in prostate cancer progression and chemoprevention regimens in an autochthonous mouse model. Clin Cancer Res 2005;11:147-53. [PubMed]
- Bjørnland K, Winberg JO, Odegaard OT, et al. S100A4 involvement in metastasis: deregulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in osteosarcoma cells transfected with an anti-S100A4 ribozyme. Cancer Res 1999;59:4702-8. [PubMed]
- Du M, Wang G, Ismail TM, et al. S100P dissociates myosin IIA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J Biol Chem 2012;287:15330-44. [Crossref] [PubMed]
- Buckley NE, D'Costa Z, Kaminska M, et al. S100A2 is a BRCA1/p63 coregulated tumour suppressor gene with roles in the regulation of mutant p53 stability. Cell Death Dis 2014;5:e1070. [Crossref] [PubMed]
- Naz S, Ranganathan P, Bodapati P, et al. Regulation of S100A2 expression by TGF-beta-induced MEK/ERK signalling and its role in cell migration/invasion. Biochem J 2012;447:81-91. [Crossref] [PubMed]