
Long noncoding RNA MAFG-AS1 facilitates bladder cancer tumorigenesis via regulation of miR-143-3p/SERPINE1 axis
Introduction
Bladder cancer is the ninth most common carcinoma worldwide, with more than 430,000 patients diagnosed with each year (1). Transitional cell carcinoma is the main histological type, accounting for over 90% of bladder cancer cases. For early and mid-stage bladder cancer patients, surgery is the optimal treatment is suitable, frustratingly, it is prone to relapse. For patient with metastases, palliative treatments such as radiotherapy and chemotherapy are the most common treatment strategy (2). Although the diagnosis and treatment of bladder cancer have been made significant advances, the 5-year survival for invasive patients are only 50%, furthermore, the recurrence rate of bladder cancer is 15–90% within 5 years (2,3). Therefore, understanding the molecular mechanisms of bladder cancer progression will help to identify possible targets for cancer therapy.
Long noncoding RNAs (lncRNAs) are transcripts that are >200 nucleotides long (4). Although they do not code proteins, they can regulate gene expression at the transcriptional and post-transcriptional levels and epigenetic modifications (5). An increasing number of studies have demonstrated that lncRNAs participate in the tumorigenesis and progression of numerous cancers, which been the focus of anti-tumour therapy (6-8). For example, lncRNA LNMAT1 contributes to lymphatic metastasis by epigenetic activation of CCL2 in bladder cancer (9). Chen et al. (10) reported that lncRNA OXCT1-AS1 acts as an miR-455-5p sponge to inhibit its binding to JAK1, leading to elevated JAK1 expression and enhanced bladder cancer aggressiveness. Dai et al. (11) demonstrated that lnc-MUC20-9 impeded bladder cancer cell growth, migration and invasion via suppression of ROCK1. However, the functions and underlying mechanisms of lncRNAs in bladder cancer progression remain unclear.
Aberrant expression of the novel lncRNA MAFG antisense RNA 1 (MAFG-AS1) was recently identified in a variety of cancers and facilitated cancer progression (12-16). However, its role and underlying mechanisms remain unclear in bladder cancer. Basing on the TCGA database analysis, MAFG-AS1 was found upregulation in bladder cancer tissues, more importantly, patients with high expression of MAFG-AS1 showed poor overall survival. It implied that MAFG-AS1 play an importance role in bladder cancer.
In present research, we demonstrated that MAFG-AS1 promotes bladder cancer cell proliferation, migration and invasion via competitive inhibition of miR-143-3p and elevation of SERPIN1 levels. Our findings provide a novel insight into tumorigenesis and reveal a promising therapeutic target for bladder cancer.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1971).
Methods
Clinic samples
A total of 52 pairs of bladder cancer tissue samples and corresponding adjacent normal tissues were obtained from Xiangya Hospital, Central South University from Oct 2018 to Oct 2019. All patients enrolled in this research didn’t receive any preoperative therapy. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the institutional ethics committee of Xiangya Hospital, Central South University (approval No. 2019032542). Informed consent has been signed by all patients before specimen collection. The pathological features were acquired from patients’ medical records. Specimen were fast frozen in liquid nitrogen and kept at −80 °C.
Cell culture and transfection
Human bladder cancer cell lines (J82, 5637, UM-UC-3 and T24) and normal cell SV-HUC-1 were obtained from COBIOER (Nanjing, China). J82 and UM-UC-3 were maintained in Minimum Eagle’s medium containing 10% FBS. 5637 and T24 were maintained in RPMI1640 supplemented with 10% FBS. SV-HUC-1 were cultured in F-12K medium containing 10% FBS.
SiRNAs targeting the lncRNA MAFG-AS1 sequences were purchased from GenePharma (Shanghai, China). The sequences information as shown in Table 1. Scramble oligonucleotides were used as a negative control. MiR-143-3p mimics and inhibitor were obtained from GeneCopoeia (Guangzhou, China). SERPINE1 cDNA ORF plasmid was purchased from Sino Biological (Beijing, China). Cells were transfected by Lipofectamine 2000 (Thermo Fisher, USA) following the manufacturers’ instructions. Forty-eight h later, cells were collected for subsequent experiments.
Table 1
| Name | Sequence |
|---|---|
| siRNAs | |
| MAFG-AS1 siRNA-1 | GGGCAAUUCCAACCAAGAAAC |
| MAFG-AS1 siRNA-2 | GCUGCAGUGAGCUGUGAUCAU |
| Scramble siRNA | UUCUCCGAACGUGUCACGU |
| qRT-PCR | |
| MAFG-AS1 forward | GGGACGGAGACAAATGACGG |
| MAFG-AS1 reverse | GCAGGCTCCCTGACACGTA |
| SERPINE1 forward | GAGGATGAAAGAAACAGCCAGCT |
| SERPINE1 reverse | CCCGCTATGAAATTAGATTCACGT |
| GAPDH forward | ACACCCACTCCTCCACCTTT |
| GAPDH reverse | TTACTCCTTGGAGGCCATGT |
qRT-PCR assay
RNA isolation was performed using TRIzol reagent (Thermo Fisher, USA). cDNA was synthesized using the GoScriptTM Kit (Promega, USA) following the product’s instructions. qRT-PCR analysis was conducted on LightCycler480 system (Roche, Germany). GAPDH and U6 used as internal control for normalizing. Each reaction was conducted in triplicate. The primer sequences were listed in Table 1.
MTT and colony formation assay
For cell proliferation analysis, approximately 5×103 cells were grown in 96-well plates and maintained for 24 h. Then, each well was added 20 µL MTT solution and maintained at 37 °C for 4 h. After removing the supernatant, each well was replenished by 150 µL DMSO and maintained at 37 °C for 10 min. Finally, the absorbance was examined at 570 nm with a microplate reader (Molecular Devices, USA).
For colony formation experiment, 1×103 cells were seeded into a 35 mm dish and maintained for two weeks. Next, cells were fixed with 4% paraformaldehyde (PFA) and stained by 0.01% crystal violet dye.
Wound healing and invasion assay
For wound healing experiment, cells were cultured in a 12-well plate for 24 h to reach nearly 90% confluence. Cell wounds were scratched using a sterile pipette tip. Then, cells were washed by PBS 3 times and cultured with serum-free medium. The wound was photographed at 0 and 24 h.
For invasion assay, 1×105 cells suspended in serum-free medium were added into the upper chamber coated with Matrigel (Corning, USA), while medium containing 20% FBS was added into the lower chambers. After maintaining at 37 °C for 24 h, the non-invading cells were cleaned by cotton swabs, and the invasive cells were fixed by 4% PFA and stained in 0.1% crystal violet for 15 min. Finally, the invasive cells were captured under the microscope (Nikon, Japan).
Cell apoptosis assay
Cells were collected and centrifuged at 2,000 rpm for 5 min. Then cells were incubated with Annexin V-FITC and PI (BioLegend, USA) according to manufacture protocol. After staining at room temperature in the dark for 20 min, the suspending cells was added with 500 µL Binding buffer. Finally, cells apoptosis was analysed by flow cytometry (CytoFlex, Beckman, USA) within 1 hour.
In vivo animal experiment
Four-to-five-week-old BALB/c nude mice were purchase from Hunan SJA Laboratory Animals Center of the Chinese Academy of Sciences (Changsha, China) and fed according to the guidelines authorized by the Animal Care Committee of the Xiangya Hospital, Central South University. One ×106 cells suspended in 0.2 mL PBS were subcutaneously injected into the right flank of animals. Tumor growth was measured and recorded every 5 days. Twenty-five days later, animals were sacrificed after anesthetized and stripped the tumors. Tumor volume was calculated by the formula: (length × width2)/2.
Subcellular fractionation
The nuclear and cytosolic fractions of 5637 and T24 cells were isolated with PARIS Kit (Thermo Fisher, USA) following to the manufacturer’s protocol.
Luciferase reporter analysis
The fragment of MAFG-AS1 and SERPINE1 comprising miR-143-3p binding sites were obtained by PCR and cloned into a psiCHECK-2 vector to construct wild-type psiCKECK2-MAFG-AS1 (Wt-MAFG-AS1) and psiCKECK2-SERPINE1 (Wt-SERPINE1) or mutant psiCKECK2-MAFG-AS1 (Mut-MAFG-AS1) and psiCKECK2-SERPINE1 (Mut-SERPINE1) vectors. 5637 and T24 cells were respectively transfected with Wt or Mut recombinant plasmids, together with miR-NC or miR-143-3p mimics. Forty-eight hours later, the luciferase activity was determined with the Dual-Luciferase reporter assay Kit (Promega, Madison, USA) following the manufacturer's instructions.
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the EZ-Magna RIP kit (Millipore, USA) according to the protocol. Simply, cells were lysed in RIP lysis buffer containing proteinase and RNase inhibitor. The cell lysate was centrifuged after 10 min of incubation. Then, the cell supernatant extract was incubated magnetic beads pre-conjugated with AGO2 antibody (Millipore, USA) or IgG at 4 °C overnight. RNA was purified from immunoprecipitation complex and qRT-PCR detection was used to evaluate the level of MAFG-AS1, miR-143-3p and SERPINE1.
Statistical analysis
All experiments were repeated three times, and the data were presented by mean ± standard deviation (SD). The statistical analysis was conducted by SPSS 22.0 (SPSS Inc., IL, USA). Student’s t-test was applied to analyse the differences between two groups. Chi-square test was performed to evaluate the relationship between MAFG-AS1 expression and clinicopathological features of bladder cancer. P<0.05 was considered statistically significant.
Results
LncRNA MAFG-AS1 was overexpressed in bladder cancer tissues and implied the poor survival
Firstly, qRT-PCR was used to examined MAFG-AS1 expression in 52 paired primary bladder cancer specimen and matched adjacent normal tissues. The result displayed that MAFG-AS1 was obviously upregulated in bladder cancer samples (Figure 1A). Moreover, MAFG-AS1 expression in lymph node (LN)-metastatic tissues was obviously higher than nonmetastatic tumor tissues (Figure 1B). The cutoff value was set basing on the median value of MAFG-AS1 expression in bladder cancer, the patients were divided into the low expression group (n=25) and the high expression group (n=27) (Figure 1C). The correlation of MAFG-AS1 expression and pathological characteristics were evaluated. The result displayed showed that the high expression of MAFG-AS1 was obviously associated with histological grade (P=0.026), TNM stage (P=0.012), and LN metastasis (P=0.047) (Table 2). Besides, MAFG-AS1 overexpressed in bladder cancer tissues was confirmed by GEPIA database (http://gepia.cancer-pku.cn) (Figure 1D). Additionally, high MEST expression indicated the poor overall survival of patients with bladder cancer from Kaplan-Meier plotter (http://kmplot.com/analysis/) (Figure 1E). These data demonstrated that MAFG-AS1 was upregulated in bladder cancer and implied the poor survival.
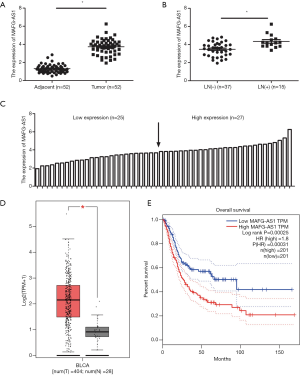
Table 2
| Features | Number of cases | MAFG-AS1 expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Age (years) | 0.500 | |||
| >60 | 26 | 14 | 12 | |
| ≤60 | 26 | 13 | 13 | |
| Gender | 0.584 | |||
| Male | 33 | 17 | 16 | |
| Female | 19 | 10 | 9 | |
| Tumor size | 0.508 | |||
| >3 cm | 28 | 15 | 13 | |
| ≤3 cm | 24 | 12 | 12 | |
| Histological grade | 0.026* | |||
| Low | 25 | 9 | 16 | |
| High | 27 | 18 | 9 | |
| TNM grade | 0.012* | |||
| I–II | 28 | 10 | 18 | |
| III–IV | 24 | 17 | 7 | |
| Lymph nodes metastasis | 0.047* | |||
| No | 37 | 16 | 21 | |
| Yes | 15 | 11 | 4 | |
LncRNA MAFG-AS1 facilitated bladder cancer cell proliferation, migration and invasion
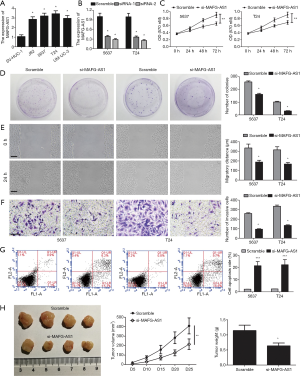
To determine the role of MAFG-AS1 in bladder cancer, firstly, MAFG-AS1 expression in bladder cancer cells (J82, 5637, T24 and UM-UC-3) and normal uroepithelial cell SV-HUC-1 was analyzed. We observed that MAFG-AS1 were markedly upregulated in tumor cell lines compared with SV-HUC-1 (Figure 2A). Next, MAFG-AS1 silencing in 5637 and T24 cells were performed via transfecting MAFG-AS1 siRNAs. The result demonstrated that MAFG-AS1 was obviously downregulated in 5637 and T24 cells (Figure 2B). MTT and clone formation assays showed that MAFG-AS1 silencing significantly inhibited cell proliferation and growth compare with the control group (Figure 2C,D). Wound scratch and invasion assays demonstrated that MAFG-AS1 silencing significantly impeded cell migration and invasion (Figure 2E,F). Flow cytometer detection showed that MAFG-AS1 depletion markedly increased cell apoptosis (Figure 2G). Moreover, MAFG-AS1 depletion significantly suppressed the tumor growth in vivo (Figure 2H). These findings suggested that MAFG-AS1 act as an oncogene in bladder cancer progression.
MiR-143-3p targeted MAFG-AS1
In order to uncover the mechanism of MAFG-AS1 on bladder cancer tumorigenesis, we firstly investigated its subcellular location. We observed that MAFG-AS1 was preferentially enriched in the cytoplasm (Figure 3A). This result implied that MAFG-AS1 could competitively interact with cytoplasmic miRNAs. So, the potential miRNA targets of MAFG-AS1 were predicted using starBase v3.0. MiR-143-3p was a predicted target and was chosen for verification as it has been reported to be involved in oncogenesis and cancer progression (17-19). The wild type and mutant type of MAFG-AS1 containing miR-143-3p binding site were constructed as showing in Figure 3B. The luciferase activity assay was conducted in 5367 and T24 cells. The result displayed that co-transfection of miR-143-3p mimics and Wt-MAFG-AS1 markedly reduced the luciferase activity (Figure 3C). Additionally, RIP experiment was performed to examine the interaction between miR-143-3p and MAFG-AS1. MAFG-AS1 and miR-143-3p were significantly enriched in the AGO2 immunoprecipitate comparing with the IgG control group (Figure 3D).
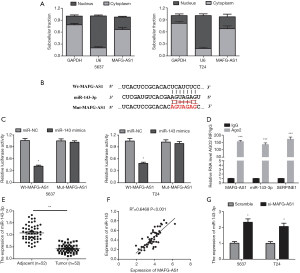
Next, the expression of miR-143-3p level in the 52 pairs of bladder cancer samples and matched adjacent normal tissues was analyzed. MiR-143-3p was found downregulation and negatively correlated with MAFG-AS1 expression in bladder cancer samples (Figure 3E,F). Besides, we observed that MAFG-AS1 silencing could evidently elevate miR-143-3p level in 5367 and T24 cells (Figure 3G). Collectively, our result confirmed that miR-143-3p was a target of MAFG-AS1.
SERPINE1 was a target of MiR-143-3p
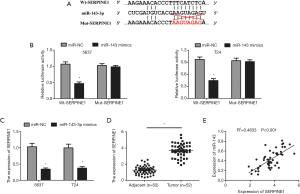
Three online softs TargetScan, miRDB and microRNA were performed to identify the possible target genes of miR-143-3p. We found that serpin family member 1 gene (SERPINE1) (also named PAI1) was a candidate target gene (Figure 4A). In order to confirm it, luciferase reporter analysis was executed. The result displayed that miR-143-3p mimics depressed the luciferase activity of Wt-SERPINE1 but not in the Mut-SERPINE1 (Figure 4B). Moreover, the expression of SERPIN1 was markedly reduced in 5367 and T24 cells transfected with miR-143-3p mimics (Figure 4C). Notably, RIP assay found that miR-143-3p and SERPINE1 were prominently enriched in the AGO2 immunoprecipitate (Figure 3D). Additionally, in bladder cancer tissues, qRT-PCR displayed that SERPINE1 was obviously upregulated and negatively correlated with that of miR-143-3p (Figure 4D,E). Collectively, our results indicated that SERPINE1 was a target of miR-143-3p.
MAFG-AS1 facilitated bladder cells tumorigenesis by regulating miR-143-3p/SERPINE1 axis
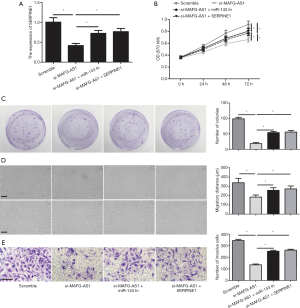
Whether miR-143-3p or SERPIN1 participated in promotive effect on bladder cancer malignant phenotypes mediated by MAFG-AS1, rescue assays were executed. As shown by qRT-PCR, miR-143-3p inhibition evidently reduce the MAFG-AS1 depletion-induced suppressing effect on the SERPIN1 expression. Meanwhile, SERPINE1 overexpression alleviated MAFG-AS1 depletion-mediated downregulation in the SERPINE1 expression (Figure 5A). Moreover, we observed that MAFG-AS1 depletion-impeded cells proliferation and growth were obviously attenuated by miR-143-3p inhibitors or SERPINE1 overexpression (Figure 5B,C). Wound scratch and Transwell invasion assay showed that suppressed cell migration and invasion mediated by MAFG-AS1 silencing were significantly alleviated by miR-143-3p inhibition or SERPINE1 overexpression (Figure 5D,E). Our finding indicated that MAFG-AS1 played its pro-oncogenic roles in bladder cancer via regulating miR-143-3p-SERPINE1 axis.
Discussion
Studies have shown that numerous lncRNAs are involved in tumorigenesis and have the potential to be used as biomarker in bladder cancer (6-8). For example, lncRNA OIP5-AS1 was considered as a useful biomarker for clinical progression and poor prognosis (19). LncRNA ATB promoted bladder cancer cell proliferation, migration, and invasion by regulating miR-126, and it was regarded as a potential prognostic biomarker and therapeutic target for bladder cancer (20). TUG1 facilitates bladder cancer cell growth and chemoresistance by modulating CCND2 via EZH2-associated silencing of miR-194-5p, which suggested that TUG1 can be used as a novel therapeutic target and biomarker for bladder cancer (21). It can be seen that lncRNAs have crucial role in the development of bladder cancer, and it is worthwhile to focus on the function and underlying mechanism of lncRNAs. The present study found that MAFG-AS1 was upregulated in bladder cancer samples and cell lines. MAFG-AS1 knockdown suppressed bladder cancer cell proliferation, migration and invasion. We investigated the mechanisms involved and found that MAFG-AS1 exerted its pro-oncogenic roles via competitive inhibition of miR-143-3p and elevation of SERPIN1 levels. Overall, our study revealed the role and mechanism of the MAFG-AS1/miR-143-3p-SERPINE1 axis in bladder cancer progression.
MAFG-AS1 was reported to be upregulated and play an oncogene role in several common carcinomas, such as colorectal cancer, breast cancer, lung cancer, hepatocellular carcinoma and gastric cancer (12-16); however, its role in bladder cancer remains unclear. We observed that MAFG-AS1 was upregulated in bladder cancer tissue samples and cell lines. Further analysis revealed that MAFG-AS1 expression was positively correlated with histological grade, TNM stage and LN metastasis. Moreover, patients with high expression of MAFG-AS1 showed reduced survival. As confirmed by a series of cell function experiments in vitro, MAFG-AS1 silencing markedly suppressed bladder cancer cell malignant phenotypes. In agreement with previous reports in other carcinomas, these results confirmed that MAFG-AS1 plays an oncogene role in bladder cancer.
Many studies have reported that lncRNAs perform their roles by acting as miRNA sponges and de-suppressing their targeting mRNAs (19). Thus, the lncRNA/miRNA/mRNA signal axis presents the most canonical model (22). For example, lncRNA HOTAI contributes to angiogenesis by modulating the miR-126/SCEL axis during burn wound healing (23). It was reported that lncRNA XIST promotes thyroid cancer cell proliferation and tumour growth by sponging miR-34a to regulate MET expression (24). Zhao et al. (25) demonstrated that lncRNA TUSC8 suppressed breast cancer progression by regulating the miR-190b-5p/MYLIP axis. In the present study, we confirmed that MAFG-AS1 performed its functions by sponging miR-143-3p to upregulate SERPINE1 levels. It was reported that miR-143-3p acts as a tumour suppressor in various carcinomas; for example, miR-143-3p impeded osteosarcoma progression by repressing stemness cell properties via targeting pro-oncogene KIA1429 (17). In breast cancer, miR-143-3p suppressed cell proliferation and invasion by targeting pyruvate carboxylase (18). In bladder cancer, miR-143-3p also was confirmed to be a tumour suppressor as it could inhibit cell growth by modulating Akt and ERK5 (26,27). Moreover, lncRNA FOXD2-AS1 enhanced gemcitabine resistance via sponging miR-143 in bladder cancer (22). It is known that miRNAs may have more than one target gene (28), and we found that SERPINE1 was the target gene of miR-143-3p. Moreover, the miR-143-3p/SERPIN1 axis was involved in bladder cancer progression.
SERPINE1 is a serine protease inhibitor that is mainly involved in suppression of tissue and urokinase-type plasminogen activators (29). SERPINE1 and urokinase-type plasminogen activator participated in tumour metastasis, as it proteolytically degraded the extracellular matrix (29). Numerous studies have reported that SERPINE1 dysregulation played an oncogenic role in various carcinomas; for example, SERPINE1 promoted cell proliferation, migration and invasion by modulating epithelial-mesenchymal transition in gastric cancer (29). Arroyo-Solera et al. revealed that SERPINE1 overexpression increased cell proliferation and promoted invasiveness and metastasis in head and neck squamous cell carcinoma (30). In osteosarcoma, SERPINE1 was shown to facilitate cell invasion and metastasis by upregulating MMP-13 (31). In bladder cancer, overexpression of SERPINE1 was closely correlated with bladder urothelial carcinoma aggressiveness and/or poor survival, but also contributed to cell invasiveness (32). In the present study, we revealed that SERPINE1 was overexpressed in bladder cancer tissues and acted as a target gene of miR-143-3p. We also showed that SERPINE1 is involved in the modulation of MAFG-AS1 in bladder cancer proliferation, migration and invasion.
Taken together, our results indicate that MAFG-AS1 is overexpressed in bladder cancer. MAFG-AS1 silencing inhibited bladder cancer cell proliferation, migration and invasion by modulating the miR-143-SERPINE1 axis. Our findings provide a novel insight into tumorigenesis and reveal a promising therapeutic target for bladder cancer.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1971
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1971
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1971). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethics committee of Xiangya Hospital, Central South University (approval No. 2019032542), and written informed consents were obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fujii Y. Prediction models for progression of non-muscle-invasive bladder cancer: A review. Int J Urol 2018;25:212-8. [Crossref] [PubMed]
- Qiu F, Zhang MR, Zhou Z, et al. lncRNA MIR503HG functioned as a tumor suppressor and inhibited cell proliferation, metastasis and epithelial-mesenchymal transition in bladder cancer. J Cell Biochem 2019;120:10821-9. [Crossref] [PubMed]
- Oughton JB, Poad H, Twiddy M, et al. Radical cystectomy (bladder removal) against intravesical BCG immunotherapy for high-risk non-muscle invasive bladder cancer (BRAVO): a protocol for a randomised controlled feasibility study. BMJ Open 2017;7:e017913. [Crossref] [PubMed]
- Yu C, Longfei L, Long W, et al. LncRNA PVT1 regulates VEGFC through inhibiting miR-128 in bladder cancer cells. J Cell Physiol 2019;234:1346-53. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- He A, He S, Peng D, et al. Prognostic value of long non-coding RNA signatures in bladder cancer. Aging (Albany NY) 2019;11:6237-51. [Crossref] [PubMed]
- Jiang L, Li Z, Wang R. Long noncoding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications Int J Oncol 2019;55:585-96. (Review). [PubMed]
- Luo L, Wang M, Li X, et al. Long non-coding RNA LOC285194 in cancer. Clin Chim Acta 2020;502:1-8. [Crossref] [PubMed]
- Chen C, He W, Huang J, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun 2018;9:3826. [Crossref] [PubMed]
- Chen JB, Zhu YW, Guo X, et al. Microarray expression profiles analysis revealed lncRNA OXCT1-AS1 promoted bladder cancer cell aggressiveness via miR-455-5p/JAK1 signaling. J Cell Physiol 2019;234:13592-601. [Crossref] [PubMed]
- Dai R, Zhou Y, Chen Z, et al. Lnc-MUC20-9 binds to ROCK1 and functions as a tumor suppressor in bladder cancer. J Cell Biochem 2020;121:4214-25. [Crossref] [PubMed]
- Cui S, Yang X, Zhang L, et al. LncRNA MAFG-AS1 promotes the progression of colorectal cancer by sponging miR-147b and activation of NDUFA4. Biochem Biophys Res Commun 2018;506:251-8. [Crossref] [PubMed]
- Li C, Wu R, Xing Y. MAFG-AS1 is a novel clinical biomarker for clinical progression and unfavorable prognosis in gastric cancer. Cell Cycle 2020;19:601-9. [Crossref] [PubMed]
- Li H, Zhang GY, Pan CH, et al. LncRNA MAFG-AS1 promotes the aggressiveness of breast carcinoma through regulating miR-339-5p/MMP15. Eur Rev Med Pharmacol Sci 2019;23:2838-46. [PubMed]
- Ouyang H, Zhang L, Xie Z, et al. Long noncoding RNA MAFG-AS1 promotes proliferation, migration and invasion of hepatocellular carcinoma cells through downregulation of miR-6852. Exp Ther Med 2019;18:2547-53. [Crossref] [PubMed]
- Sui Y, Lin G, Zheng Y, et al. LncRNA MAFG-AS1 boosts the proliferation of lung adenocarcinoma cells via regulating miR-744-5p/MAFG axis. Eur J Pharmacol 2019;859:172465. [Crossref] [PubMed]
- Han Q, Yang J, Yang H, et al. KIAA1429 promotes osteosarcoma progression by promoting stem cell properties and is regulated by miR-143-3p. Cell Cycle 2020;19:1172-85. [Crossref] [PubMed]
- Pinweha P, Phillips CA, Gregory PA, et al. MicroRNA-143-3p targets pyruvate carboxylase expression and controls proliferation and migration of MDA-MB-231cells. Arch Biochem Biophys 2019;677:108169. [Crossref] [PubMed]
- Yang J, Jiang B, Hai J, et al. Long noncoding RNA opa-interacting protein 5 antisense transcript 1 promotes proliferation and invasion through elevating integrin alpha6 expression by sponging miR-143-3p in cervical cancer. J Cell Biochem 2019;120:907-16. [Crossref] [PubMed]
- Zhai X, Xu W. Long Noncoding RNA ATB Promotes Proliferation, Migration, and Invasion in Bladder Cancer by Suppressing MicroRNA-126. Oncol Res 2018;26:1063-72. [Crossref] [PubMed]
- Yu G, Zhou H, Yao W, et al. lncRNA TUG1 Promotes Cisplatin Resistance by Regulating CCND2 via Epigenetically Silencing miR-194-5p in Bladder Cancer. Mol Ther Nucleic Acids 2019;16:257-71. [Crossref] [PubMed]
- An Q, Zhou L, Xu N. Long noncoding RNA FOXD2-AS1 accelerates the gemcitabine-resistance of bladder cancer by sponging miR-143. Biomed Pharmacother 2018;103:415-20. [Crossref] [PubMed]
- Jiang B, Tang Y, Wang H, et al. Down-regulation of long non-coding RNA HOTAIR promotes angiogenesis via regulating miR-126/SCEL pathways in burn wound healing. Cell Death Dis 2020;11:61. [Crossref] [PubMed]
- Liu H, Deng H, Zhao Y, et al. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res 2018;37:279. [Crossref] [PubMed]
- Zhao L, Zhou Y, Zhao Y, et al. Long non-coding RNA TUSC8 inhibits breast cancer growth and metastasis via miR-190b-5p/MYLIP axis. Aging (Albany NY) 2020;12:2974-91. [Crossref] [PubMed]
- Noguchi S, Mori T, Hoshino Y, et al. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett 2011;307:211-20. [Crossref] [PubMed]
- Noguchi S, Yasui Y, Iwasaki J, et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett 2013;328:353-61. [Crossref] [PubMed]
- An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med 2017;21:185-92. [Crossref] [PubMed]
- Yang JD, Ma L, Zhu Z. SERPINE1 as a cancer-promoting gene in gastric adenocarcinoma: facilitates tumour cell proliferation, migration, and invasion by regulating EMT. J Chemother 2019;31:408-18. [Crossref] [PubMed]
- Arroyo-Solera I, Pavon MA, Leon X, et al. Effect of serpinE1 overexpression on the primary tumor and lymph node, and lung metastases in head and neck squamous cell carcinoma. Head Neck 2019;41:429-39. [PubMed]
- Hirahata M, Osaki M, Kanda Y, et al. PAI-1, a target gene of miR-143, regulates invasion and metastasis by upregulating MMP-13 expression of human osteosarcoma. Cancer Med 2016;5:892-902. [Crossref] [PubMed]
- Li X, Dong P, Wei W, et al. Overexpression of CEP72 Promotes Bladder Urothelial Carcinoma Cell Aggressiveness via Epigenetic CREB-Mediated Induction of SERPINE1. Am J Pathol 2019;189:1284-97. [Crossref] [PubMed]


