
Screening of potential genes and transcription factors involved in post-radiation cognitive dysfunction in mice via bioinformatics
Introduction
Brain radiotherapy has been an indispensable mainstay treatment for patients with primary brain tumors or brain metastases originating from extracranial tumors (1-5). Long-term tumor control or cure, slowing tumor growth, and killing metastatic cells palliatively or prophylactically are achieved by various modes of brain radiotherapy (6,7). However, radiotherapy has been shown to cause adverse effects on the central nervous system (CNS), including both acute and delayed complications. Furthermore, cancer patients may experience a wide range of neurotoxic symptoms, including problems in learning and memory, executive function impairment, loss of motor coordination, and intellectual decline, which have been long recognized as major problems in long-term survivors in the pediatric population (8,9). Radiotherapy-induced cognitive decline drastically reduces the patients’ quality of life, which draws oncologists’ close attention (10). Despite rapidly growing concerns about radiation-induced cognitive disability, the metabolic and cellular mechanisms underlying such damage remain poorly understood. Moreover, there are neither effective preventive measures nor long-term treatments available (11). Recent studies in brain cancer patients and rodent models on radiation-related neurofunctional sequelae showed that various anatomical changes occurred in the non-tumor tissue during irradiation. The affected zones included the hippocampus (Hipp) and subventricular zone (SVZ) of the brain lateral ventricles (12). A hypothesis has been proposed that early changes that are not detectable on the gross anatomical level, including vascular damages, oligodendrocyte loss, demyelination, neuroinflammation, and radiation-induced brain injury, could synergize over time to generate long-term macrostructural and microstructural abnormalities that in turn could result in permanent cognitive decline (13-15). Although several markers have been identified to predict the damage and cognitive decline in delayed complications, few markers of acute complications have been found (5,9). Therefore, there is a strong need to study the molecular mechanisms of these complications and identify novel therapeutic targets that would help delay or ameliorate postradiation cognitive dysfunction. Moreover, the early identification of potential biomarkers and understanding their molecular mechanisms of action in postradiation cognitive dysfunction continues to be debated and requires further exploration and research. In our study, we aimed to analyze potential genes and transcription factors to explore the possible mechanisms of cognitive dysfunction in the early postradiation period using bioinformatics analysis.
Methods
Microarray data
Published microarray data on mRNA expression profiles were reanalyzed to explore possible mechanisms of cognitive dysfunction in the early postradiation period. The GSE115735 mRNA microarray dataset was retrieved and downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), a public functional genomics data repository. Microarray data were obtained from GPL23038 Clariom S Assay Platform (Affymetrix, Inc., Santa Clara, CA, USA) and included gene expression data corresponding to three Hipp and three brain posterior lateral ventricle (pLV) tissues from mice with postradiation cognitive dysfunction and control mice, respectively. Previous studies showed that neurogenesis mainly occurs in two brain regions: the subgranular zone (SGZ) in the hippocampal dentate gyrus and the SVZ (16,17). The three Hipp and three pLV tissues included the SGZ and SVZ samples, respectively, which enabled us to analyze the abnormal transcriptional activity related to postradiation cognitive dysfunction. These mice received a total dose of 10 Gy in 2 fractions (2 × 5 Gy). The control animals were littermates who were handled similarly but did not receive radiation. Cognition in mice was assessed through their performance in the novel object recognition task with a long habituation phase using odorless objects that do not retain any olfactory cues on days 40–44. A discrimination index was used to assess the difference between the two groups quantitatively. Then, brain tissue was collected at day 50 post radiation (18,19).
Differential expression analysis
Differentially expressed mRNAs (DEMs) between postradiation cognitive dysfunction and normal control samples were identified using bioinformatics analysis. Normalized gene expression data were downloaded as original raw data to be used for further analysis, and this dataset contained the expression information of 28,847 mRNAs. The differential expression analysis was performed using a Student’s t-test in the Limma R package (20). Only mRNAs with false discovery rate (FDR)-values of <0.05 and logFC > |mean (abs [logFC]) + 2*sd (abs [logFC])| were considered as DEMs. Venn diagrams were plotted using an online tool, VENNY 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Protein-protein interaction network construction
Protein-protein interaction (PPI) network construction was performed to screen for potential biomarkers. We used Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) (21-23), which is an online software tool that provides critical assessment and integration of PPIs, including direct (physical) and indirect (functional) interactions derived from computational predictions, knowledge transfer between organisms, and interactions aggregated from additional (primary) databases. Based on the STRING analysis, only the known interactions that were proven by biological experiments with a combined score of >0.15 were retrieved as significant items for further analysis. To visualize PPI networks, the Cytoscape software, which is a powerful analytical tool for providing a unified conceptual framework by the integration of biomolecular interaction networks, was used (24). The top three hub genes were predicted using cytoHubba.
Functional enrichment pathway analysis
Gene Ontology (GO) analysis was conducted to assess differentially expressed genes between the irradiated mouse brain and control brain tissues at the functional level (25). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed to further explore the cellular and molecular functions of the differentially expressed genes (26). ClusterProfiler R package was used to carry out the analysis and visualization of GO and KEGG (27). We selected the top significantly enriched items and confirmed their correlations with genomic differences by a thorough search in PubMed.
Prediction of transcription factors and construction of Protein-Transcription Factor (P-TF) Network
Transcription factors (TFs) involved in the regulation of potential target genes were identified. The iRegulon, Cytoscape plugin (http://iregulon.aertslab.org) was used to predict TF-target genes. Normalized Enrichment Score (NES) is the maximum enrichment score of the TF. NES > 2 was set as the cutoff criterion to screen for potential TFs. Network analysis was conducted between the DEMs and these potential TFs.
Statistical analysis
Part of the statistical analysis has been done by the bioinformatic tools mentioned above. The R software (version 3.4.1) was used for all the rest of statistical analyses. Differential expression levels of mRNA were estimated by a two-tailed Student’s t-test. Fisher’s test was used to identify the significant GO terms and KEGG pathways. Spearman correlation coefficients were calculated to evaluate the correlations. A P-value <0.05 was considered statistically significant.
Results
Identification of DEMs
Based on the Limma R package, a total of 64 and 84 mRNAs were identified as differentially expressed between the irradiated mice brain and control brain tissues in the Hipp and pLV regions, respectively. Fourteen genes were differentially expressed in both tissues: AGPAT2, STOM, CD53, UBE2C, OIP5, GPD2, DEPDC1, CTSS, CD40, PTPN22, SIGLEC11, OTUD7B, EPHX1, and RBL1. Thresholds of FDR < 0.05 and 0.65 (due to the limited DEMs) served as the cutoff values of log2FC (Figure 1).
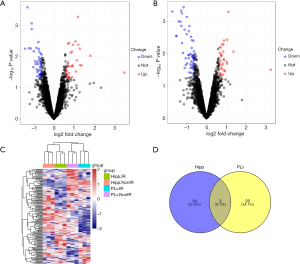
PPI network construction
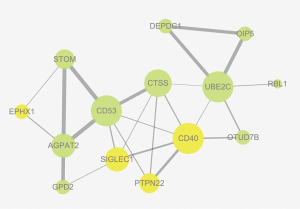
With the help of the STRING online tool, a total of 24 PPI pairs were identified with the combined score of >0.15. Among these, the top three hub nodes were CTSS, CD53, and CD40 (Figure 2).
Functional and pathway enrichment analysis
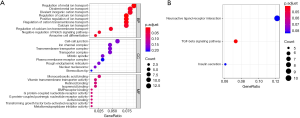
GO functional analysis of potential target genes revealed 415 categories associated with biological processes, 175 cell component-associated categories, and 437 functional GO molecular function-associated categories forming top 10, 9, and 10 categories, respectively (Figure 3A). A total of 257 signaling pathways were identified (P<0.05). KEGG functional analysis revealed 39 categories, and the top three categories are shown in Figure 3B.
Prediction of TFs and P-TF network construction
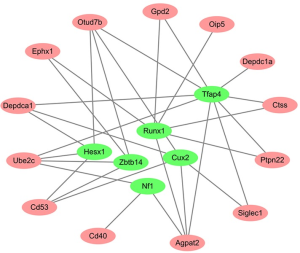
Six TFs associated with the regulation of target genes were identified (NES > 2): TFAP4, RUNX1, CUX2, ZBTB14, NF1, and HESX1. The P-TF network was constructed as shown in Figure 4.
Discussion
Our analysis identified 14 differentially expressed genes between the irradiated mouse brain and control brain tissues. Based on the PPI network analysis, the top three hub genes were CTSS, CD40, and CD53. The top three TFs predicted to be associated with target gene regulation were TFAP4, RUNX1, and CUX2. Functional and pathway enrichment analysis (GO and KEGG) revealed that these genes might play a role in the pathogenesis of postradiation cognitive dysfunction in the early period (<1 month) after brain radiotherapy.
Cognitive dysfunction has become an increasingly important issue for patients with brain tumors and brain metastases due to the wide use of brain radiotherapy, especially the whole-brain radiation therapy (28). Brain radiotherapy has harmful effects on multiple organ systems, such as the CNS, which lead to neurotoxic adverse reactions, including acute and delayed complications. A phase III trial was terminated early because the patients assigned to brain radiotherapy were more likely to have a deterioration in learning and memory function despite an improved intracranial control (29). An analysis of long-term survivors found that radiation-induced neurocognitive decline started at approximately four months after treatment initiation, which was followed by an improvement and then a progressive irreversible deterioration in neurocognitive function at 12 months or later after irradiation (30). However, it is important to note that tumor progression during this time period could also adversely affect cognition. Therefore, research in the subtle early manifestations of irradiation damage to the CNS during the early period (<1 month) is essential. Several studies demonstrated that permanent cognitive disability may result from the early synergism of CNS damages leading to macrostructural and microstructural alterations (5,9,13). In the current study, 14 differentially-expressed genes associated with postradiation cognitive dysfunction during the early period after irradiation were identified in both the Hipp and pLV brain areas: AGPAT2, STOM, CD53, UBE2C, OIP5, GPD2, DEPDC1, CTSS, CD40, PTPN22, SIGLEC11, OTUD7B, EPHX1, and RBL1. These genes were obtained to identify the functional significance of DEMs. Furthermore, the PPI network analysis showed that CTSS, CD53, and CD4 were the hub nodes in the PPI network built in this study.
Several researchers have speculated that the early dysfunction of the surviving neuronal cells alters the signaling microenvironment, thus influencing progenitor cell differentiation and cognitive capacity over the long term (9,31,32). KEGG pathway analysis in our study suggested that the neuroactive ligand-receptor interaction pathways may play important roles in early CNS damages caused by radiation. A recent study indicated that α-synuclein could cause miRNA deregulation, which targeted neuroactive ligand-receptor interaction pathways in the early stage of Parkinson's disease in the drosophila model (33). GO classification and enrichment analysis showed that the regulation of calcium-ion transport and mitotic spindle associated pathway may play an important role. The regulation of calcium-ion transport is closely related to mitochondrial energy metabolism, and recent studies demonstrated that alterations in calcium homeostasis were at the basis of increased vulnerability of neurons during aging-related processes, such as cognitive decline and synaptic dysfunctions (34). Radiation is now known to suppress both the proliferation of hippocampal SGZ progenitor cells and their differentiation into neurons (9). Neurogenesis in the murine hippocampus was reduced after 5 Gy or 10 Gy of intracranial X-ray irradiation, which was associated with impaired spatial learning (35,36).
In our study, TFs were predicted to establish biological mechanisms that contributed to postradiation cognitive dysfunction. The top three TFs including TFAP4, RUNX1, and CUX2 were involved in potential target gene regulation. TFAP4, TF activating enhancer binding protein 4, is a TF that is selectively expressed in the brain, and it may play an important role in cellular damage compensation through neurobehavioral recovery and neurovascular remodeling after brain injury (37,38). The potential biological mechanisms of TFAP4 involve endothelial cell proliferation, cell survival and migration, as well as neuroprotection or improved neurovascular coupling through the regulation of kinase insert domain receptor, a vascular endothelial growth factor receptor (39,40). Furthermore, TFAP4 was shown to be involved in transcriptional repression in Alzheimer's disease and postoperative cognitive dysfunction (41,42). In recent years, the role of runt-related TF 1 (RUNX1) in neural development has been studied in different models. These studies suggested that RUNX1 played an important role in the proliferation and differentiation of neural progenitor cells (NPCs), the control of neurite outgrowth, and in the axonal pathfinding (43,44). In the hippocampus, which is a major site of postnatal/adult neurogenesis, NPCs generate newborn dentate gyrus granule cell neurons throughout life, and this process of hippocampal neurogenesis is thought to be critical for the normal hippocampal function (45). Animal studies have elucidated the pathological effects of radiation on hippocampal progenitor cell biology. Research using these models has demonstrated that exposure to therapeutic doses of irradiation resulted in increased apoptosis, decreased cell proliferation, and decreased neuronal differentiation in the neurogenic region of the hippocampus (46). Moreover, RUNX1 has been suggested to play a role in peripheral and CNS development, in defining different brain compartments, and in consolidation of the specific neuronal identity in the developing mouse nervous system (47,48). CUX2 is a member of the homeodomain TF family containing cut repeat DNA-binding sequences and is primarily expressed in nervous tissues. Reports have shown that CUX2 might act as a trigger to end expansion and exit the cell cycle (49,50). However, further studies are required to understand how CUX2 influences the balance between the decision to reenter or exit the cell cycle and to fully evaluate how these CUX2 functions might play a role in early postradiation cognitive dysfunction. These TFs have close associations with neurovascular repair, nerve cell proliferation, differentiation, and neural development and may be key regulators during postradiation cognitive dysfunction development, which confirms that the current study method is effective in identifying key TFs. Results of key TF predictions suggest that AP4 and RUNX1 play pivotal roles in the regulation of potentially important key genes, such as CTSS, that contribute to postradiation cognitive dysfunction.
Conclusions
Multifactorial origins contribute to the irreversible cognitive deterioration after intracranial radiotherapy because the CNS contains many structures that are potentially sensitive to radiation (51). In our study, 14 DEMs were identified as gene products involved in postradiation cognitive dysfunction based on bioinformatics analysis. CTSS, CD40, and CD53 were the top three hub genes based on the PPI network construction, and TFAP4, RUNX1, CUX2 were the top three TFs predicted. GO enrichment, KEGG pathway analysis, and previous studies indicated that these genes might be associated with the pathogenesis of postradiation cognitive dysfunction in the early period. All these conclusions provide new insights into the roles of crucial TFs associated with postradiation cognitive dysfunction in mouse models. However, no other reports at present have shown that these TFs were linked with early postradiation cognitive dysfunction. Therefore, additional studies on the association of these potential biomarkers with postradiation cognitive dysfunction are required to further substantiate the results obtained in our research.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1767). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chi A, Komaki R. Treatment of brain metastasis from lung cancer. Cancers 2010;2:2100-2137. [Crossref] [PubMed]
- Shi L, Olson J, D'Agostino R Jr, et al. Aging masks detection of radiation-induced brain injury. Brain Res 2011;1385:307-316. [Crossref] [PubMed]
- Greene-Schloesser D, Moore E, Robbins ME. Molecular pathways: radiation-induced cognitive impairment. Clin Cancer Res 2013;19:2294-2300. [Crossref] [PubMed]
- Moore ED, Kooshki M, Wheeler KT, et al. Differential expression of Homer1a in the hippocampus and cortex likely plays a role in radiation-induced brain injury. Radiat Res 2014;181:21-32. [Crossref] [PubMed]
- Wu PH, Coultrap S, Pinnix C, et al. Radiation induces acute alterations in neuronal function. PLoS One 2012;7:e37677. [Crossref] [PubMed]
- Bilimagga RS, Nirmala S, Rishi KS, et al. Role of palliative radiotherapy in brain metastases. Indian J Palliat Care 2009;15:71. [Crossref] [PubMed]
- Wadasadawala T, Gupta S, Bagul V, et al. Brain metastases from breast cancer: management approach. J Cancer Res Ther 2007;3:157. [Crossref] [PubMed]
- Davis CM, Roma PG, Armour E, et al. Effects of X-ray radiation on complex visual discrimination learning and social recognition memory in rats. PLoS One 2014;9:e104393. [Crossref] [PubMed]
- Greene-Schloesser D, Robbins ME, Peiffer AM, et al. Radiation-induced brain injury: A review. Front Oncol 2012;2:73. [Crossref] [PubMed]
- Gustafsson M, Edvardsson T, Ahlström G. The relationship between function, quality of life and coping in patients with low-grade gliomas. Support Care Cancer 2006;14:1205-12. [Crossref] [PubMed]
- Lee YW, Cho HJ, Lee WH, et al. Whole brain radiation-induced cognitive impairment: pathophysiological mechanisms and therapeutic targets. Biomol Ther (Seoul) 2012;20:357. [Crossref] [PubMed]
- Makale MT, McDonald CR, Hattangadi-Gluth JA, et al. Mechanisms of radiotherapy-associated cognitive disability in patients with brain tumours. Nat Rev Neurol 2017;13:52. [Crossref] [PubMed]
- Attia A, Page BR, Lesser GJ, et al. Treatment of radiation-induced cognitive decline. Curr Treat Options Oncol 2014;15:539-50. [Crossref] [PubMed]
- Palmer TD, Willhoite AR, Gage FH. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol 2000;425:479-94. [Crossref] [PubMed]
- Peiffer AM, Leyrer CM, Greene-Schloesser DM, et al. Neuroanatomical target theory as a predictive model for radiation-induced cognitive decline. Neurology 2013;80:747-53. [Crossref] [PubMed]
- Hellström NAK, Björk-Eriksson T, Blomgren K, et al. Differential recovery of neural stem cells in the subventricular zone and dentate gyrus after ionizing radiation. Stem Cells 2009;27:634-41. [Crossref] [PubMed]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 2011;70:687-702. [Crossref] [PubMed]
- Soria B, Martin-Montalvo A, Aguilera Y, et al. Human Mesenchymal Stem Cells Prevent Neurological Complications of Radiotherapy. Front Cell Neurosci 2019;13:204. [Crossref] [PubMed]
- Leger M, Quiedeville A, Bouet V, et al. Object recognition test in mice. Nat Protoc 2013;8:2531-7. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Franceschini A, Lin J, von Mering C, et al. SVD-phy: improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics 2016;32:1085-7. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res 2006;34:D322-6. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. [Crossref] [PubMed]
- Brown PD, Ahluwalia MS, Khan OH, et al. Whole-brain radiotherapy for brain metastases: evolution or revolution? J Clin Oncol 2018;36:483. [Crossref] [PubMed]
- Chang EL, Wefel JS, Hess KR, et al. Neurocognition in patients with brain metastases treated with radiosurgery or radiosurgery plus whole-brain irradiation: a randomised controlled trial. Lancet oncol 2009;10:1037-44. [Crossref] [PubMed]
- McTyre E, Scott J, Chinnaiyan P. Whole brain radiotherapy for brain metastasis. Surg Neurol Int 2013;4:S236. [Crossref] [PubMed]
- Snyder JS, Kee N, Wojtowicz JM. Effects of adult neurogenesis on synaptic plasticity in the rat dentate gyrus. J Neurophysiol 2001;85:2423-31. [Crossref] [PubMed]
- Wong CS, Van der Kogel AJ. Mechanisms of radiation injury to the central nervous system: implications for neuroprotection. Mol Interv 2004;4:273. [Crossref] [PubMed]
- Kong Y, Liang X, Liu L, et al. High throughput sequencing identifies microRNAs mediating α-synuclein toxicity by targeting neuroactive-ligand receptor interaction pathway in early stage of drosophila Parkinson's disease model. PLoS One 2015;10:e0137432. [Crossref] [PubMed]
- Navakkode S, Liu C, Soong TW. Altered function of neuronal L-type calcium channels in ageing and neuroinflammation: Implications in age-related synaptic dysfunction and cognitive decline. Ageing Res Rev 2018;42:86-99. [Crossref] [PubMed]
- Mizumatsu S, Monje ML, Morhardt DR, et al. Extreme sensitivity of adult neurogenesis to low doses of X-irradiation. Cancer Res 2003;63:4021-7. [PubMed]
- Tada E, Parent JM, Lowenstein DH, et al. X-irradiation causes a prolonged reduction in cell proliferation in the dentate gyrus of adult rats. Neuroscience 2000;99:33-41. [Crossref] [PubMed]
- Dzietko M, Derugin N, Wendland MF, et al. Delayed VEGF treatment enhances angiogenesis and recovery after neonatal focal rodent stroke. Transl Stroke Res 2013;4:189-200. [Crossref] [PubMed]
- Shimotake J, Derugin N, Wendland M, et al. Vascular endothelial growth factor receptor-2 inhibition promotes cell death and limits endothelial cell proliferation in a neonatal rodent model of stroke. Stroke 2010;41:343-9. [Crossref] [PubMed]
- Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med 2003;9:669-76. [Crossref] [PubMed]
- Ahmad S, Hewett PW, Wang P, et al. Direct evidence for endothelial vascular endothelial growth factor receptor-1 function in nitric oxide–mediated angiogenesis. Circ Res 2006;99:715-22. [Crossref] [PubMed]
- Wang Y, Huang A, Gan L, et al. Screening of potential genes and transcription factors of postoperative cognitive dysfunction via bioinformatics methods. Med Sci Monit 2018;24:503. [Crossref] [PubMed]
- Zhou F, Yao H, Wu JY, et al. Activation of Group II/III metabotropic glutamate receptors attenuates LPS-induced astroglial neurotoxicity via promoting glutamate uptake. J Neurosci Res 2006;84:268-77. [Crossref] [PubMed]
- Theriault FM, Nuthall HN, Dong Z, et al. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J Neurosci 2005;25:2050-61. [Crossref] [PubMed]
- Yoshikawa M, Masuda T, Kobayashi A, et al. Runx1 contributes to the functional switching of bone morphogenetic protein 4 (BMP4) from neurite outgrowth promoting to suppressing in dorsal root ganglion. Mol Cell Neurosci 2016;72:114-22. [Crossref] [PubMed]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell 2008;132:645-60. [Crossref] [PubMed]
- Monje ML, Mizumatsu S, Fike JR, et al. Irradiation induces neural precursor-cell dysfunction. Nat Med 2002;8:955-62. [Crossref] [PubMed]
- Levanon D, Brenner O, Negreanu V, et al. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech Dev 2001;109:413-7. [Crossref] [PubMed]
- Stifani N, Freitas ARO, Liakhovitskaia A, et al. Suppression of interneuron programs and maintenance of selected spinal motor neuron fates by the transcription factor AML1/Runx1. P Natl A Sci 2008;105:6451-6.
- Capaldo E, Iulianella A. Cux2 serves as a novel lineage marker of granule cell layer neurons from the rhombic lip in mouse and chick embryos. Dev Dyn 2016;245:881-96. [Crossref] [PubMed]
- Yamada M, Clark J, McClelland C, et al. C ux2 activity defines a subpopulation of perinatal neurogenic progenitors in the hippocampus. Hippocampus 2015;25:253-67. [Crossref] [PubMed]
- Connor M, Karunamuni R, McDonald C, et al. Dose-dependent white matter damage after brain radiotherapy. Radiother Oncol 2016;121:209-16. [Crossref] [PubMed]


