
OSMR and SEPT9: promising biomarkers for detection of colorectal cancer based on blood-based tests
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers, and its incidence rates are still rapidly increasing in several areas with an aging population and increasing changes in lifestyle habits (1). Taking precaution through CRC screening and the removal of precancerous polyps could dramatically decrease incidence rates (2,3). Several screening options such as colonoscopy, fecal occult blood tests (FOBTs), air contrast barium enemas and CT virtual endoscopy could help to alleviate the incidence of CRC (2,4-6). Colonoscopy is an effective method for early detection, but it has a limited application due to its expensive cost, painful colonic procedure and risk of intestinal perforation (7). FOBT is still a commonly used auxiliary diagnostic method worldwide. However, it has low sensitivity and poor specificity and cannot be used for screening high-risk groups (2). Air contrast barium enema and CT virtual endoscopy are vulnerable to the influence of the intestinal cleaning state. The former has the advantages of the inspection of the whole colon, lacking biopsy or polyps; the latter is costly and requires specialized equipment and techniques. Both methods are not widely accepted by patients (4). Thus, a non-damaging, successful and simple method for screening is needed.
Cancer-specific DNA methylation is common and has been shown to be an early event in several cancer types (8,9). DNA methylation, mRNA and microRNA can be analyzed in stool samples (10-13). However, the transportation and processing of samples can be very inconvenient. The detection and monitoring of circulating tumor-derived DNA in the cancer patient plasma offer exciting opportunities for tumor treatment (14,15). Changes in DNA methylation levels, variations in cancer-associated copy number, and gene mutations have been found in the plasma of patients with different cancer types (16,17). Most of these studies on plasma DNA as a cancer marker have focused on the detection of specific molecular targets by PCR-based methods, such as shotgun massively parallel sequencing (MPS), fluorescence-based real-time PCR assay (MethyLight) and digital PCR (18-21).
In the present study, we analyzed OSMR, which is methylated frequently in CRC and rarely in cancers of non-digestive organs (22). We also analyzed SEPT9 as a reference to select the best candidates indicative of CRC for early diagnosis and monitoring of markers in plasma samples (23-26). SEPT9 is an existing biomarker for detecting CRC in blood and is commercially available from the Epigenomics Company. In this study, we aimed to evaluate the effectiveness in CRC diagnosis by analyzing the methylation of SEPT9 and OSMR in plasma, and serum biomarkers (Carcinoembryonic antigen: CEA; carbohydrate antigen: CA19-9, CA72-4 and CA242) were also analyzed in parallel. These serum biomarkers are currently used in routine early diagnosis and for monitoring tumor markers for gastrointestinal neoplasm in our hospital.
Methods
Ethics
The study was approved by the ethics committee of Peking University Cancer Hospital & Institute. Written informed consent was obtained from all participants for obtaining blood or tissue samples.
Human CRC plasma and tissue specimens
Plasma specimens, including 187 patients with CRC (100 males and 87 females, mean age 63.8 years), 25 polyp patients (17 males and 8 females, mean age 50.6 years) and 109 healthy controls (46 males and 63 females, mean age 60.8 years), were gathered from November 2011 to April 2013. About 40 paired tissues matching these plasma were collected immediately after operation and stored in a −80 °C freezer. Inflammatory bowel disease, familial adenomatous polyposis and hereditary non-polyposis CRC were not included in this study. Approximately 109 healthy adults were eligible controls recruited from employee physical examination at Peking University Cancer Hospital &Institute. The control cancer histories were checked in past medical examination records and the subjects provided informed consent for the detection of their methylation state. Plasma samples of polyp patients were obtained from the Sixth Affiliated Hospital, Sun Yat-sen University, and collected before colonoscopy. Plasma samples of CRC patients were collected before surgical resection. None of the patients with CRC received chemotherapy, radiotherapy or any surgical intervention.
Five-milliliter peripheral blood samples were collected using EDTA-containing tubes before any therapeutic intervention at the Department of Gastrointestinal Surgery, Peking University of Cancer Hospital & Institute. The tumor tissues of the CRC patients were obtained during their cancer resection surgeries.
RNA isolation, quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Real-time PCR was performed using the ABI 7500 Fast Real-Time PCR System (Life Technologies, Carlsbad, California, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The primer sequences used are listed in Table 1.
Full table
DNA extraction in tissue and plasma samples
Total DNA of frozen tissue was extracted by a TIANamp genomic DNA kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The DNA was eluted in 50 µL elution buffer, and its concentration was measured with a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware, USA) and finally stored in a −80 °C freezer.
Plasma preparation was performed on all of the peripheral blood samples by centrifugation at 2,000 rpm for 10 min at 4 °C, and the plasma portion was recentrifugated at 12,000 rpm for 10 min at 4 °C. Plasma samples were stored at −80°C until needed. Cell-free DNA molecules from 2 mL plasma were extracted according to the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, California, USA). During isolation of plasma DNA, carrier RNA (Qiagen, Valencia, California, USA) was added to increase the production.
DNA bisulfite treatment and MethyLight analysis of SEPT9 and OSMR
Sodium bisulfite conversion of tissue and plasma DNA was performed using the EZ DNA methylation-gold kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer’s protocol. Subsequently, 1 µL of the eluted tissue and 3 µL of plasma DNA were used for each respective MethyLight PCR reaction.
Bisulfite-treated DNA was analyzed by a fluorescence-based real-time PCR assay, which was described previously as MethyLight. One pair of primers and a probe for each gene (OSMR, SEPT9 and COL2A1) were designed specifically to bind to the bisulfite-converted DNA (Table 1). The COL2A1 gene was analyzed to control DNA amplification and normalized for input DNA. Specificity of the reactions for methylated DNA was confirmed by separately amplifying positively methylated cell line RKO DNA and unmethylated human control DNA with each set of primers and probes. PCRs were carried out in 20 µL volumes containing 1× PCR buffer, 5 mM MgCl2, 250 µM dNTP mixture, 0.5 µM of each primer, 0.3 µM of each probe, 1 or 3 µL bisulfite-treated tissue or plasma DNA, and 0.05 U/µL Taq DNA polymerase (HotStar Taq plus, Qiagen, Valencia, California, USA). PCR was carried out in the ABI 7500 fast (Life Technologies, Carlsbad, CA, USA) using the following conditions: 95 °C for 5 min, followed by 55 cycles of 94 °C for 30 s, 60 °C for 60 s. The Ct values of each gene were normalized to the Ct values of COL2A1.
Carcinoembryonic antigen (CEA), CA199, CA72.4 and CA242 test
All blood samples for the CEA, CA19-9, CA72-4 and CA242 tests were analyzed in the clinical laboratory of Peking University Cancer Hospital. Serum levels of CEA, CA199, CA72.4 and CA242 that were higher than 5.0, 37, 6.7 and 20 µg/L, respectively, were considered to be in the pathological range.
Statistical analysis
Statistical analysis was carried out by Statistical Analysis System (8.1; SAS Institute, Cary, NC, USA) and Graphpad Prism 5.0 (GraphPad software, San Diego, CA, USA). The non-parametric t test was used to calculate the difference in bisulfite-converted DNA between paired tissue samples. The Wilcoxon matched-pairs test was used to analyze the differences in expression levels between paired tissue samples. The χ2 test was used to explore associations between methylation and serum biomarker levels and clinicopathological features. The selection of cut-off values was based on the receiver operating characteristics (ROC) curve. P<0.05 was taken as the statistical significance level.
Results
SEPT9 and OSMR mRNA expression in CRC tissues and their methylation levels in CRC and polyp tissues
We compared SEPT9 expression in 30 paired CRC tissue samples and their adjacent non-cancer tissue samples by qPCR. SEPT9 was found to be down-regulated significantly compared with their adjacent non-cancer tissues (P=0.006). Then SEPT9 methylation (mSEPT9) was analyzed in 30 paired CRC tissue samples and 15 paired polyp tissue samples. There was a significantly higher mSEPT9 level in tumors than in adjacent non-cancer tissue samples (P<0.0001), and the methylation levels in polyp tissue samples were also higher than those of the normal counterparts (P<0.0001) (Figure 1A).
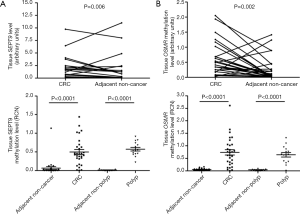
We analyzed OSMR expression level in 30 paired CRC tumors and their adjacent non-cancer tissues by quantitative real-time PCR (qPCR). OSMR was found to be down-regulated significantly in primary tumor specimens compared with their adjacent non-cancer tissues (P=0.002). OSMR methylation (mOSMR) was analyzed in 30 paired CRC tissue samples and 15 paired polyp tissue samples. mOSMR level was significantly higher in tumors than those in adjacent non-cancer tissue (P<0.0001). Methylation levels in polyp tissue samples were also higher than in normal tissues (P<0.0001) (Figure 1B).
Sensitivity of blood-based tumor markers for CRC and polyp patient detection
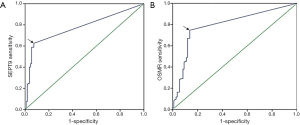
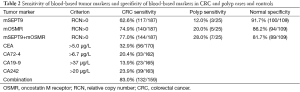
The ROC curve based on the methylation level in plasma of CRC patients and normal controls had an area under the curve value of 0.777 for SEPT9 and 0.796 for OSMR, respectively (Figure 2). As shown in Table 2, mSEPT9 was positive in 117 (62.6%) patients with CRC, 3 (12.0%) patients with polyps, and the specificity in healthy controls was 100 (91.8%) healthy controls using the 1/3 analysis method. mOSMR was positive in 140 (74.9%) patients with CRC, 5 (20.0%) patients with polyps, and the specificity was 94 (86.2%) healthy controls using the 2/3 analysis method. A combination of mSEPT9 and mOSMR improved sensitivity to 77.0% but decreased true-negatives to 81.7% (89/109) in CRC patients.
Full table
Among these 221 CRC patients, CEA, CA72.4, CA199 and CA242 were positive in 56 (32.9%), 33 (20.4%), 23 (13.9%) and 39 (23.9%) patients respectively. The combination of methylation and serum markers dramatically improved sensitivity to 83.0% in CRC patients (Table 2).
Association of biomarkers in blood with clinicopathological factors
The associations of mSEPT9 and mOSMR in plasma, serum biomarkers or their combination with clinicopathological features are shown in Table 3. When we compared mSEPT9 and mOSMR level in colon and rectal cancers, we did not find a significant difference between the two groups (for mSEPT9, P=0.097 and for mOSMR, P=0.378). There was also no significant difference for serum biomarkers between the colon and rectal cancer groups (P>0.05). mSEPT9 was 57.1% and 69.5% positive for stage I+II and III+IV CRC cases (P=0.083), respectively. mOSMR was 74.4% and 76.8% positive for stage I+II and III+IV CRC cases (P=0.689), respectively. There were significant differences (P=0.030, 0.001, 0.044 and 0.009, for CEA, CA199, CA72.4 and CA242, respectively) between the stage I+II and III+IV patients with positive CEA, CA199 CA72.4 and CA242 levels, respectively. A combination of methylation and serum markers improved stage I+II and III+IV detection to 81.6% and 84.7% (P=0.603), respectively.
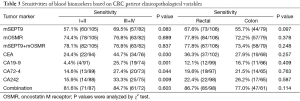
Full table
Comparisons of mSEPT9 and mOSMR levels in plasma and their matched tumors
We selected 40 CRC tissue samples that expressed mSEPT9 and mOSMR and collected their matched plasma to analyze the correlation in the methylation level of these two markers. SEPT9 methylation levels were 87.5% vs. 70.0% in CRC tissues and their matched plasma, and the levels for mOSMR were 95.0% vs. 75.0%. A correlation between the methylation levels in tumor tissues and plasma was present in SEPT9 (P=0.001) but not in OSMR (P=0.058). Unmethylated SEPT9 and OSMR were observed in 12 and 10 plasma samples, respectively. No false-positive was observed in these two markers in plasma samples (Table 4).

Full table
Discussion
To reduce the mortality of CRC, CRC screening is needed to identify tumors at earlier stages. However, current screening methods for clinical application are either invasive (i.e., colonoscopy) or have low sensitivity and specificity (i.e., CEA, CA199 and FOBT). With a case-control study, mSEPT9 in plasma was consistently demonstrated to be useful for detecting CRC in several clinical studies in several countries (America, Germany, China and Korea) (25,27,28). The CRC detection rates with mSEPT9 ranged from 36.6% to 90% with different methods (11,16,17,24,25,29). Therefore, we believe that a panel of blood-based methylation tests will be valuable in screening.
In the present study, mSEPT9 and mOSMR were selected for evaluation as noninvasive plasma screening markers for CRC in Chinese CRC patients. mSEPT9 is a familiar marker that was developed by the Epigenomics company (25,30,31). The overall sensitivity in our study was 62.6%, and the overall specificity was 91.7%. The sensitivity and specificity of mSEPT9 were similar to the previous reports. mOSMR has been reported to be frequently methylated in CRC tissues (90%) and less frequently methylated in gastric (33%) and pancreatic (20%) cancers, but rarely methylated in cancers of other organs (13). The overall sensitivity in our study was 74.9%, and the overall specificity was 86.2%. In contrast to our results in Chinese patient plasma, the sensitivity of fecal mOSMR in previous studies was 38% by TaqMan-MSP (32). The evaluation of mOSMR in CRC plasma shows that the sensitivity is higher than that in fecal samples.
To demonstrate the levels of mSEPT9 and mOSMR in Chinese patients, we detected mSEPT9 and mOSMR in CRC tissues and their matched adjacent non-cancer tissues. mSEPT9 and mOSMR were highly detected in cancer tissues compared with their matched adjacent non-cancer tissues (both P<0.0001). Therefore, the levels of mSEPT9 in Chinese patients were higher in cancer tissues than their matched adjacent non-cancer tissues, but the sensitivity of plasma mSEPT9 was higher than that in Koreans (36.6%), likely due to the different detection methods (33). In addition, the level of mOSMR in Chinese patients was the same as in the previous report by Deng et al. (22), but the sensitivity of plasma mOSMR was higher in our current investigation due to the method of detection or ethnic variations in the methylation pattern of OSMR.
Notably, false positive methylation of SEPT9 and OSMR was not detected in plasma, and our findings suggested that mSEPT9 and mOSMR in plasma could match their methylation in the corresponding tumor tissues. Although the P value of OSMR failed to reach statistical significance (P=0.058), it may be induced by minute quantity of circular tumor DNA and exhibit less sensitivity with current qPCR (34).
We also analyzed the relationship between the level of mSEPT9 or mOSMR in plasma samples and the level of serum biomarkers and clinicopathologic characteristics. There was not a significant difference in mSEPT9 or mOSMR in CRC tumors and plasma samples of patients with CRC with different locations and TNM stages. In daily practice, serum CEA, CA72-4, CA19-9 and CA242 concentrations are now used for screening and follow-up examination in CRC patients. We analyzed pretreatment serum CEA, CA72-4, CA19-9 and CA242 concentrations in our study subjects. The sensitivity of these serum biomarkers (CEA: 32.9%, CA72.4: 20.4%, CA199: 13.9% and CA242: 23.9%) were lower compared with mSEPT9 (62.6%) and mOSMR (74.9%). In our study, the sensitivity of the combined analysis of mSEPT9 and mOSMR improved to 77.0% in CRCs, and the specificity of combined analysis did not decrease substantially when compared with the single marker (Table 3). However, the combination of methylation and serum biomarkers dramatically improved CRC detection (83.0%). Thus, multiple markers, including plasma mSEPT9 and mOSMR, and serum CEA, CA72-4, CA19-9 and CA242, might be more useful for CRCs in screening and monitoring.
In conclusion, plasma mSEPT9 and mOSMR were detected at a high frequency in Chinese patients. Both were not correlated with clinicopathologic characteristics. Our investigation assessed the level of mSEPT9 and mOSMR, and clinical serum biomarkers in CRCs. The results indicate that mSEPT9 and mOSMR are sensitive biomarkers for the detection of CRC, with sensitivities of 62.6% and 74.9%, especially for the sensitivity of combination (77.0%). Therefore, the combination of multiple markers might need to be validated in future investigation (in our study, the sensitivity was 83.0% for the combination of methylation and serum biomarkers). Both of these markers are more sensitive than FOBT and serum biomarkers commonly used in clinics. Therefore, the analyses of methylation levels of SEPT9 and OSMR in plasma, especially OSMR as a new biomarker, are useful for the diagnosis of CRC. The panel including SEPT9, OSMR and serum biomarker (CEA, CA19-9, CA72-4 and CA242) analysis appears feasible and convenient for the diagnosis of CRC.
Acknowledgments
We thank Guoshuang Feng (from Chinese Center for Disease Control and Prevention) for his kind help in data statistics.
Funding: This work was supported by Beijing Natural Science Foundation of China (No. 7133231), Peking University Cancer Hospital Research Fund (No. 2013 independent 7), The National High Technology Research and Development Program of China (863 Program, No. 2014AA020603).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.03.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Peking University Cancer Hospital & Institute. Written informed consent was obtained from all participants for obtaining blood or tissue samples.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Center MM, Jemal A, Smith RA, et al. Worldwide variations in colorectal cancer. CA Cancer J Clin 2009;59:366-78. [Crossref] [PubMed]
- Edwards BK, Noone AM, Mariotto AB, et al. Annual Report to the Nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer 2014;120:1290-314. [Crossref] [PubMed]
- Winawer SJ. The multidisciplinary management of gastrointestinal cancer. Colorectal cancer screening. Best Pract Res Clin Gastroenterol 2007;21:1031-48. [Crossref] [PubMed]
- Cotton PB, Durkalski VL, Pineau BC, et al. Computed tomographic colonography (virtual colonoscopy): a multicenter comparison with standard colonoscopy for detection of colorectal neoplasia. JAMA 2004;291:1713-9. [Crossref] [PubMed]
- Rockey DC, Paulson E, Niedzwiecki D, et al. Analysis of air contrast barium enema, computed tomographic colonography, and colonoscopy: prospective comparison. Lancet 2005;365:305-11. [Crossref] [PubMed]
- Thakkar K, El-Serag HB, Mattek N, et al. Complications of pediatric colonoscopy: a five-year multicenter experience. Clin Gastroenterol Hepatol 2008;6:515-20. [Crossref] [PubMed]
- Okines A, Verheij M, Allum W, et al. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21:v50-4. [Crossref] [PubMed]
- Kwon HJ, Kim JH, Bae JM, et al. DNA methylation changes in ex-adenoma carcinoma of the large intestine. Virchows Arch 2010;457:433-41. [Crossref] [PubMed]
- Nishioka Y, Ueki T, Hokazono K, et al. Comparative detection of aberrantly methylated DNA in preoperative and postoperative stool from patients with colorectal cancers. Int J Biol Markers 2015;30:e81-7. [Crossref] [PubMed]
- Alexander RJ, Raicht RF. Purification of total RNA from human stool samples. Dig Dis Sci 1998;43:2652-8. [Crossref] [PubMed]
- Wu XD, Song YC, Cao PL, et al. Detection of miR-34a and miR-34b/c in stool sample as potential screening biomarkers for noninvasive diagnosis of colorectal cancer. Med Oncol 2014;31:894. [Crossref] [PubMed]
- Wu CW, Ng SS, Dong YJ, et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 2012;61:739-45. [Crossref] [PubMed]
- Giasuddin AS, Jhuma KA, Haq AM. Applications of free circulating nucleic acids in clinical medicine: recent advances. Bangladesh Med Res Counc Bull 2008;34:26-32. [Crossref] [PubMed]
- Tong YK, Lo YM. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta 2006;363:187-96. [Crossref] [PubMed]
- Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414-23. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761-8. [Crossref] [PubMed]
- Trinh BN, Long TI, Laird PW. DNA methylation analysis by MethyLight technology. Methods 2001;25:456-62. [Crossref] [PubMed]
- He Q, Chen HY, Bai EQ, et al. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet 2010;202:1-10. [Crossref] [PubMed]
- Weisenberger DJ, Trinh BN, Campan M, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res 2008;36:4689-98. [Crossref] [PubMed]
- Deng G, Kakar S, Okudiara K, et al. Unique methylation pattern of oncostatin m receptor gene in cancers of colorectum and other digestive organs. Clin Cancer Res 2009;15:1519-26. [Crossref] [PubMed]
- Grützmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One 2008;3:e3759 [Crossref] [PubMed]
- deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009;55:1337-46. [Crossref] [PubMed]
- Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317-25. [PubMed]
- Powrózek T, Krawczyk P, Kucharczyk T, et al. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med Oncol 2014;31:917. [Crossref] [PubMed]
- Su XL, Wang YF, Li SJ, et al. High methylation of the SEPT9 gene in Chinese colorectal cancer patients. Genet Mol Res 2014;13:2513-20. [Crossref] [PubMed]
- Lee SG, Park TS, Oh SH, et al. De novo acute myeloid leukemia associated with t(11;17)(q23;q25) and MLL-SEPT9 rearrangement in an elderly patient: a case study and review of the literature. Acta Haematol 2011;126:195-8. [Crossref] [PubMed]
- Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011;9:133. [Crossref] [PubMed]
- Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 2014;60:1183-91. [Crossref] [PubMed]
- Molnár B, Tóth K, Barták BK, et al. Plasma methylated septin 9: a colorectal cancer screening marker. Expert Rev Mol Diagn 2015;15:171-84. [Crossref] [PubMed]
- Kim MS, Louwagie J, Carvalho B, et al. Promoter DNA methylation of oncostatin m receptor-beta as a novel diagnostic and therapeutic marker in colon cancer. PLoS One 2009;4:e6555 [Crossref] [PubMed]
- Lee HS, Hwang SM, Kim TS, et al. Circulating methylated septin 9 nucleic Acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol 2013;6:290-6. [Crossref] [PubMed]
- Tham C, Chew M, Soong R, et al. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer 2014;120:3131-41. [Crossref] [PubMed]


